Abstract
The present study portrays the isolation of four phenylpropanoids – ferulic acid (FA), sinapic acid (SA), caffeic acid (CA), and chlorogenic acid (CHA) from the water extract of Suaeda maritima (L.) Dumort, a phytochemically less explored Indian medicinal plant. Further, synthesis and characterization of silver and gold nanoparticles using the isolated phenylpropanoids were done. The silver nanoparticles synthesized from S. maritima water extract along with silver nano-conjugated forms of the isolated compounds exhibited appreciable anti-leukemic activity against K562 cells (human myeloid leukemia). Especially, the ferulic and CA-conjugated silver nanoparticles showed significant (P < .01) activity against leukemia.
Introduction
“Umiri” or “Mattaumiri” is botanically equated as Suaeda maritima (L.) Dumort, belongs to the family Amaranthaceae. Leaves of this plant are traditionally used for the treatment of hepatitis and it has antiviral, antibacterial, and antioxidant activities (Patra et al. Citation2011, Ravikumar et al. Citation2011). This medicinal halophyte shows the presence of biologically active secondary metabolites like polyphenols, flavonoids, alkaloids, sterols, and cardiac glycosides (El-Latif et al. Citation2014). Encapsulation of metals especially as silver and gold as nanoparticles with the plant extracts has been reported as one of the reliable methods to relatively potentiate the bioactivity and improve the efficacy (Ahmed et al. Citation2016, Al-Shmgani et al. Citation2016, Nasrabadi et al. Citation2014, Murugan et al. Citation2015, Saravanakumar et al. Citation2016, Sinha and Paul Citation2015). In the present study, four phenylpropanoids – ferulic acid (FA), caffeic acid (CA), sinapic acid (SA), and chlorogenic acid (CHA) – were isolated from the aqueous extract of S. maritima. Also S. maritima extract (SMAE) was used to synthesize gold (AuNPs) and silver (AgNPs) nanoparticles. The SMAE AgNPs, isolated phenylpropanoids, and phenylpropanoids AgNPs were tested for anti-proliferative activity against K562-leukemia cells. Further, cell viability, ROS generation, and DNA fragmentation studies were performed on the isolated compounds and their respective AgNPs to find the possible mechanism of anti-leukemic potential.
Materials and methods
Materials
Histopaque 1077, and Rhodamine B, RPMI 1640, penicillin, streptomycin, pentoxifylline (POF), N-acetyl-l-cysteine (NAC), PEG6000, doxorubicin were procured from Sigma (St. Louis, MO). Fetal bovine serum (FBS) was purchased from Gibco (Waltham, MA). MTT was purchased from Himedia, Mumbai, India. Zinc perchlorate hexahydrate, Tris–HCl, Tris buffer, Titron X-100, sodium dodecyl sulfate (SDS), phenol, chloroform, iso-amyl alcohol, ethidium bromide (EtBr), and 2-vinylpyridine were procured from Merck Ltd., SRL Pvt. Ltd., Mumbai, India. Commercially available dimethyl sulfoxide (DMSO) was procured from Himedia, Mumbai, India, and was purified by vacuum distillation over KOH. All other chemicals were from Merck Ltd and SRL Pvt. Ltd. Mumbai of the highest purity grade available.
Plant collection and extraction
Fresh leaves of S. maritima (L.) Dumort were collected from the Mangrove rich Muthupettai lagoon in Tiruvarur district, Tamil Nadu, India. The species was identified with the help of Flora of Tamil Nadu Carnatic. The voucher specimen of S. maritima (NRSM 2003) was deposited in Centre for Advanced Research in Indian System of Medicine (CARISM) and compared with the herbarium available (RHT 3698) in Rapinat Herbarium, St. Joseph’s College, Tiruchirapalli, Tamil Nadu, India. The shade dried, cleaned leaves (3.5 kg) were pulverized and soaked in water for 24 h. The aqueous extract (SMAE) thus obtained was filtered, lyophilized (15.7 g), and stored at 4 °C until further use. The obtained aqueous extract was further subjected to column chromatography for the isolation of compounds.
Isolation of phenyl-propanoids
About 15 g of aqueous extract of Sueada maritima was subjected to column chromatographic separation. Initially the crude extract was separated by using Sephadex LH 20 (Sigma Aldrich, Bangalore, India). Sephadex was initially soaked for 24 h in chloroform–methanol (30:70). The column fractions were eluted using chloroform–methanol (30:70) which yielded four fractions (SM1–SM4). Based on the TLC profile, SM3 and SM4 were mixed which contained four spots. This fraction (2 g) was then advanced to Silica gel (100–200 mesh) column chromatography. Elution was done by using chloroform:methanol combinations (5, 10, 15, 20, and 30%). Of all the fractions obtained, fractions SMF5–SMF11 yielded compound 1 (60 mg). Fractions SMF14–SMF18 lead to the formation of dull white precipitate which eventually yielded compound 2 (92 mg). Fractions SMF 26–27 yielded pale yellow colored compound 3 (38 mg). Fractions 31–37 resulted in a mild precipitation which later under lyophilization yielded compound 4 (27 mg). All the fractions were dried using rotary evaporator (Buchi® Rotavap R-210, Buchi, Mumbai, India). TLC was monitored with silica gel precoated aluminum sheets (Type 60 F254, Merck, Darmstadt, Germany) and the spots were visualized in the ultraviolet light chamber, iodine chamber, 5% MeOH–H2SO4 mixture. The structure of the isolated compounds was confirmed by using 1H NMR (Bruker-300 NMR spectrometer – chemical shifts were expressed as part per million against TMS as internal reference, Bruker BioSpin AG, Fällanden, Switzerland)
Synthesis of silver and gold nanoparticles
About 5 ml of the extract containing 5 mg of lyophilized SMAE dissolved in 5 ml deionized water and filtered through Whatman filter paper no. 41 was added to 5 ml of 1 mM silver nitrate solution and stirred for 1 h at room temperature. Within 30 min, brownish yellow color was formed indicating the formation of AgNPs. Similarly, 1 mM of HAuCl4 solution was used to prepare AuNPs. The nanoparticle solution prepared was stored for 2 d and was used for further characterizations. Likewise, 5 ml of stock solution of phenylpropanoids were mixed with 5 ml of 1 × 10−3 M AgNO3 solution. Conversion of yellowish-brown color indicates the formation of AgNPs.
Characterization of AgNPs and AuNPs
Ag and Au NPs formation were confirmed by UV–Vis spectrophotometer (Lambda 25, Perkin Elmer, Waltham, MA) and by FT-IR (spectrum 100, Perkin Elmer, Waltham, MA). Particle size analysis was done with Laser Diffractometry coupled Zeta Sizer Nano-series (ZS-90 Red, Malvern Instruments, Malvern, England). The dispersed nanoparticles were taken in polystyrene cuvette and subjected to the particle size analysis. X-ray diffraction (XRD) patterns were recorded using Rigaku Ultima III diffractometer (Rigaku, Tokyo, Japan) with Cu-Kα radiation in the 2θ range from 10° to 80°. Surface analysis of AgNPs and AuNPs was done using K-Alpha instrument (XPS K-Alpha surface analysis, Thermo Fisher Scientific, Waltham, MA). Analysis was carried out from the binding energy range of 0–1350 eV by using Aluminum X-ray as source, 400 μm spot size, 5 μm step size with the pass energy of 200 eV and a dwell time of 10. The surface morphology of the AgNPs and AuNPs was analyzed using the Field Emission Transmission Electron Microscope (FE-TEM 2701F, JEOL, Tokyo, Japan).
Anti-proliferative potential against human leukemia cells (K562)
Cell lines culture and maintenance
Human myeloid leukemia cell line, K562 (CML) was obtained from NCCS, Pune (India). Cells were maintained in RPMI-1640 media supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin under 5% CO2, and 95% humidified atmosphere at 37 °C in CO2 incubator (Eppendorf New Brunswick, Hamburg, Germany). Cells were cultured and maintained in a logarithmic growth phase until a number of cells reaches at 1.0 × 106 cells per ml. PBL were isolated from heparinized blood sample from donor by standard protocol using Histopaque 1077 gradient.
In vitro cell viability assay
The dose- and duration-dependent cytotoxicity of the plant extract, isolated molecules, and their respective silver nanoparticles on K562 cell line was quantitatively estimated by tetrazolium salt, 3-[4,5-dimethylthiazol- 2-yl]-2,5-diphenil-tetrazolium bromide (MTT) assay. Cell culture procedure has been followed as per standard protocols (Rajendran et al. Citation2016). The percentage of proliferation was calculated by using the following equation:
The concentration required for a 50% inhibition of cell viability (IC50) was determined graphically (Statistica version 5.0, STAT Inc., Boca Raton, FL)
Quantitative estimation of DNA fragmentation by diphenylamine (DPA) assay
After the treatment schedule, cells were lysed with hypotonic lysis buffer followed by centrifugation. Then perchloric acid (0.5 M) was added to the pellets containing uncut DNA (re-suspended with 200 ml of hypotonic lysis buffer), and to the other half of the supernatants containing DNA fragments. Then two volumes of a solution containing 0.088 M DPA, 98% (v/v) glacial acetic acid, 1.5% (v/v) sulfuric acid, and 0.5% (v/v) of 1.6% acetaldehyde solution were added. The samples were stored at 4 °C for 48 h. The colorimetric reaction was quantitated spectrophotometrically at 575 nm. The percentage of fragmentation was calculated as the ratio of DNA in the supernatants to the total DNA.
Intracellular ROS measurement
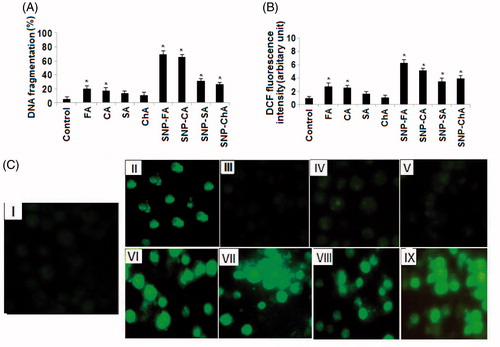
The normal PBL and K562 cell lines (2 × 105 cells per milliliter) were treated with samples for 24 h. As a positive control, those cells were incubated with H2O2 (100 μM) for 30 min in prior to the analysis. After treatment schedule, cells were washed with culture media followed by incubation with 1 μg ml−1 H2DCFDA for 30 min at 37 °C. Then the cells were washed three times with fresh culture media. DCF fluorescence was determined at 485 nm (excitation) and 520 nm (emission) by using a Hitachi F-7000 fluorescence spectrophotometer (Hitachi, Tokyo, Japan) and was also observed by fluorescence microscopy. All experiments were done in triplicate.
Results and discussion
Isolation of phenyl-propanoids
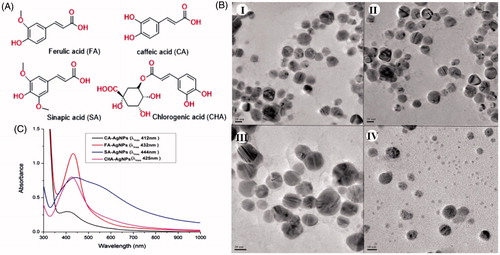
The aqueous extract of S. maritima (SMAE) was subjected to chromatographic separation(s) and four phenyl propanoids – FA, CA, SA, and CHA were isolated. The high-performance thin layer chromatography (HPTLC) (Figure S1) profile shows the presence of above four molecules as spots in the lane 1 (aqueous extract of S. maritima). The isolated molecules are depicted in the lanes 2–5, by the order of isolation. The molecules isolated were characterized using NMR spectral data and the structures were elucidated as depicted in Figure S2–S5.
Synthesis and characterization of nanoparticles
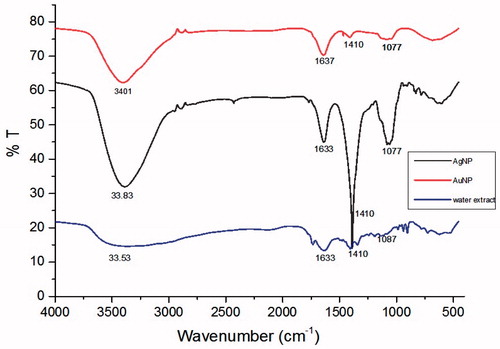
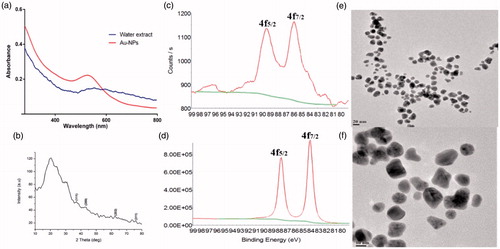
Initially, aqueous extract of S. maritima (SMAE) for its bio-reductant capacity against silver and gold salt solutions. When AgNO3 solution was mixed with the transparent SMAE solution, an immediate yellowish-brown solution was obtained, indicating the formation of AgNPs. Likewise, addition of SMAE solution to choloroauric acid (HAuCl4) solution resulted in AuNPs with pinkish red color. UV–Vis Absorption spectra of AgNPs gave a broad shoulder band from 400 nm to 450 nm () and the AuNPs had an absorption maximum at 520 nm (Elia et al. Citation2014) (). Through dynamic light scattering (DLS) experiments, AgNPs had an average particle size of 144 nm and AuNPs exhibited an average particle size of 54 nm (Figures S6 and S7). XRD analysis depicted that AgNPs showed diffraction peaks at 2θ = 38.32°, 44.47°, and 65°, corresponding to (111), (200), and (220) planes, matching the fcc silver (JCPDS file no. 04-0783) (). The average grain size of AgNPs obtained using the Debye–Scherrer formula was 26 nm. The AuNPs showed diffraction peaks at 2θ = 37.24°, 43.21°, and 62.28° corresponding to (111), (200), and (202) planes (). Further, XPS analysis performed aided in determining the chemical environment around silver and gold atoms. The Ag3d5/2 and Ag3d3/2 core level binding energies for AgNPs appear at 373 eV and 379 eV, respectively ( and ). On comparison of the binding energies of metallic silver and Nano silver, a change in the binding energy of 5 eV was observed (Figure S8B). Similarly, for AuNPs, the core level Au4f signal was observed with binding energies at 86 eV (Au4f7/2) and 90 eV (Au4f5/2), and a change in the binding energy of 2 eV was observed (Figure S9B). These shifting of XPS binding energies, when compared with parent metals (gold or silver), were in complete agreement for both AgNPs and AuNPs (Prieto et al. Citation2012, Zhou et al. Citation2010) ( and and and ). In TEM analysis, AgNPs showed spherical morphology with a particle distribution of 5–30 nm ( and ) and AuNPs were more in the 5–20 nm range ( and ) (Alam et al. Citation2015, Casaletto et al. Citation2006). In FTIR (), the broad peak at 3357 cm−1 indicates the presents of the –OH group in the plant extract. The shoulder peak at 1729 cm−1 assigned for the C = O group of carboxylic acids, aldehyde, or esters. The peak at 1633 cm−1 indicates the fingerprint region of CO, C–O, and O–H groups present in the water extract. The absorption peaks at 1401 cm−1 could be attributed to the presence of phenolic OH. The intense band at 1186 cm−1 can be assigned to the C–N stretching of amine. FTIR study indicates that the carboxyl (–C = O), hydroxyl (–OH), and amine (N–H) groups present in the water extract of S. maritima. Comparing the FT-IR spectrum of water extract with AgNP and AuNP spectrum, the broad –OH stretching peaks reduced and other absorption peaks at 1729 cm−1, 1633 cm−1 were decreased. A peak at 1401 cm−1 was found to be sharp but reduced. These observations suggest the presence of phenolic compounds in the extract (Arokiyaraj et al. Citation2016).
Figure 1. Characterization of Silver nanoparticles synthesized from plant extract: (A) UV–Vis spectrum, (B) XRD pattern, (C) XPS analysis, (D) XPS analysis of silver metal, and (E and F) HR-TEM images.
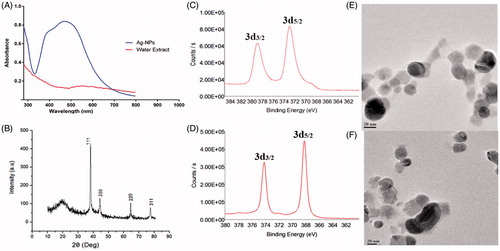
Figure 2. Characterization of gold nanoparticles synthesized from plant extract (SMAE): (A) UV–Vis spectrum, (B) XRD pattern, (C) XPS analysis, (D) XPS analysis of gold metal, and (E and F) HR-TEM images.
Since the aqueous extract of S. maritima contains predominantly four phenyl propanoids which were isolated in pure forms, AgNPs were synthesized using the four molecules (FA, CA, SA, and CHA) individually. All the four compounds formed gratifying AgNPs with AgNO3 solution. The characterization was done similar to nanoparticles obtained from SAME and thus the size and morphology nanoparticles obtained from isolated molecules were confirmed by TEM () (Singh et al. Citation2016).
Anti-proliferative potential against human leukemia cells (K562)
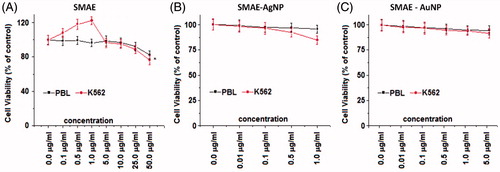
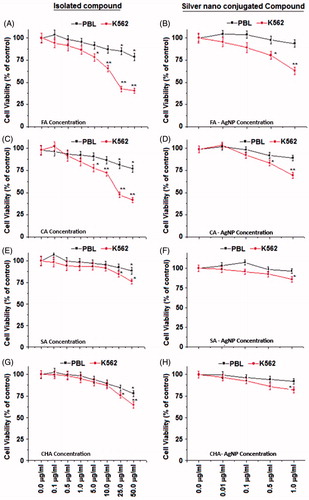
Initially, SMAE was screened for anti-proliferative effect against human chronic myeloid leukemia (K562) and human liver hepatocellular carcinoma (HepG2) cell lines, using the MTT assay. SMAE exhibited gratifying decrease in a cell viability of K562 cells at 25–50 μg/ml concentration (). The SMAE AgNPs and SMAE AuNPs were then screened for anti-proliferative activity which depicted remarkable anti-proliferative activity at a much lower concentrations (Khan et al. Citation2016). However, it was found that AgNPs from SMAE were more efficient (0.5–1.0 μg/ml), when compared with AuNPs (1.0–5.0 μg/ml) towards inhibiting the proliferation of K562 cells significantly (P < .05). When the anti-proliferative data against HepG2 cell line for SMAE, its AgNPs and AuNPs, were analyzed, the results were not gratifying. Further the phenyl propanoids and its AgNPs and AuNPs also did not exhibit promising activity of HepG2 cell lines (Figure S10A and B). Thus, AgNPs were considered for the further experiments, using the K562 cell lines. The anti-proliferative property of K562 cells were further evaluated in a concentration-dependent manner with the isolated individual phenylpropanoids. They exhibited considerable inhibition towards cell viability of K562 cells at concentrations, 25–50 μg/ml. Since, it was observed previously that AgNPs displayed remarkable anti-proliferative activity, the experiments were tried with synthesized individual phenylpropanoid AgNPs (0.1–50 μg/ml). The results were striking with the anti-leukemic evaluation of phenylpropanoid AgNPs. AgNPs from FA and CHA induced significant cell death of K562 cells at 1 μg/ml dose (. To assess the overall specificity of the AgNPs towards cytotoxicity of cells, we assessed its cytotoxic potential against normal blood cells (PBL – peripheral blood lymphocytes). Most surprisingly, we did not notice any significant (P < .05) cell death of PBL for compounds and their AgNPs at the same test concentration providing a vital scientific observation that AgNPs from molecules are specific in inhibiting the proliferation of leukemic (blood cancer cells, K562) and thus it can be inferred that the AgNPs are biologically safe.
Mechanism of action of nanoparticles against K562
With further curiosity to know the basic mechanism of this anti-leukemic action of nanoparticles, quantitative DNA fragmentation in K562 cells of isolated and nano-conjugated compounds were evaluated by using DPA assay. Our results indicate that individual FA, CA, and all four nano-conjugated forms (AgNPs) of phenylpropanoids were able to induce DNA fragmentation in K562 cells significantly (P < .05) compared with control (. Interestingly, AgNPs of FA and CA were more efficient to induce the DNA fragmentation, augmenting its anti-proliferative potential against leukemic cells. To further study the underlying mechanism causing DNA fragmentation and cell death, we studied the ROS generation by DCF2DA staining followed by spectrofluorometric analysis and fluorescence microscopy (). Significant (P < .05) ROS generations in K562 cells after administration of FA, CA, and all four phenylpropanoids AgNPs compounds indicate that the ROS induced DNA fragmentation was the cause of cell death in K562 cells (Chattopadhyay et al. Citation2015, Dash et al. Citation2015, Mahapatra et al. Citation2009).
Conclusion
In the present study, four phenylpropanoids – FA, CA, SA, and CHA were isolated from the aqueous extract of S. maritima and a green synthesis of AgNPs and AuNPs have been done using the water extract of S. maritima and its isolated phenylpropanoids. All the nanoparticles were characterized using UV–VIS, IR, TEM, XRD, and XPS studies. The S. maritima AgNPs, isolated phenylpropanoids, and respective phenylpropanoids AgNPs were tested for anti-proliferative activity against K562-leukemia cells and HepG2 cells. It was observed that AgNPs from FA and CHA displayed striking anti-cancer activities at much lower concentrations against K562 cells, without being cytotoxic towards PBL. Further, ROS generation and DNA fragmentation studies performed depicted that AgNPs from molecules fragment the DNA of K562 cells thus explaining their mechanism of anti-leukemic potential.
Sivasubramanian_et_al._supplementary_content.doc
Download MS Word (851 KB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
The authors thank the Management, SASTRA University, Thanjavur, India for the infrastructure and necessary facilities and also for the research funding through the T.R. Rajagopalan funds. The financial support by DST-SERB, Government of India under the EMR scheme (EMR/2016/000326) is also acknowledged.
References
- Ahmed Q, Gupta N, Kumar A, Nimesh S. 2016. Antibacterial efficacy of silver nanoparticles synthesized employing Terminalia arjuna bark extract. Artif Cells Nanomed Biotechnol. 1–9. doi: 10.1080/21691401.2016.1215328.
- Alam MF, Laskar AA, Zubair M, Baig U, Younus H. 2015. Immobilization of yeast alcohol dehydrogenase on polyaniline coated silver nanoparticles formed by green synthesis. J Mol Catal B: Enzym. 119:78–84.
- Al-Shmgani HSA, Mohammed WH, Sulaiman GM, Saadoon AH. 2016. Biosynthesis of silver nanoparticles from Catharanthus roseus leaf extract and assessing their antioxidant, antimicrobial, and wound-healing activities. Artif Cells Nanomed Biotechnol. 1–7. doi: 10.1080/21691401.2016.1220950.
- Arokiyaraj S, Vincent S, Saravanan M, Lee Y, Oh YK, Kim KH. 2016. Green synthesis of silver nanoparticles using Rheum palmatum root extract and their antibacterial activity against Staphylococcus aureus and Pseudomonas aeruginosa. Artif Cells Nanomed Biotechnol. 1–8. doi: 10.3109/21691401.2016.1160403. [Epub ahead of print].
- Casaletto MP, Longo A, Martorana A, Prestianni A, Venezia AM. 2006. XPS study of supported gold catalysts: the role of Au0 and Au+δ species as active sites. Surf Interface Anal. 38:215–221.
- Chattopadhyay S, Dash SK, Tripathy S, Das B, Mahapatra SK, Pramanik P, Roy S. 2015. Cobalt oxide nanoparticles induced oxidative stress linked to activation of TNF-α/caspase-8/p38-MAPK signaling in human leukemia cells. J Appl Toxicol. 35:603–613.
- Dash SK, Chattopadhyay S, Ghosh T, Dash SS, Tripathy S, Das B, et al. 2015. Self-assembled betulinic acid augments immunomodulatory activity associates with IgG response. Biomed Pharmacother. 72:144–157.
- Elia P, Zach R, Hazan S, Kolusheva S, Porat Z, Zeiri Y. 2014. Green synthesis of gold nanoparticles using plant extracts as reducing agents. Int J Nanomed. 9:4007–4021.
- El-Latif ABDRR, Mansour RMA, Sharaf M, Farag A. 2014. Three new flavonol glycosides from Suaeda maritima. J Asian Nat Prod Res. 5:434–439.
- Khan BAA, Mahapatra SK, Charanraja MR, Subramanian S, Sengan M, Narendran R, et al. 2016. Jacalin capped silver nanoparticles minimizes the dosage use of the anticancer drug, shikonin derivatives against human chronic myeloid leukemia. RSC Adv. 6:18980–18989.
- Mahapatra SK, Chakraborty SP, Majumdar S, Bag BG, Roy S. 2009. Eugenol protects nicotine-induced superoxide mediated oxidative damage in murine peritoneal macrophages in vitro. Eur J Pharmacol. 623:132–140.
- Murugan K, Sanoopa CP, Madhiyazhagan P, Dinesh D, Subramaniam J, Panneerselvam C, et al. 2015. Rapid biosynthesis of silver nanoparticles using Crotalaria verrucosa leaves against the dengue vector Aedes aegypti: what happens around? An analysis of dragonfly predatory behaviour after exposure at ultra-low doses. Nat Prod Res. 30:826–833.
- Nasrabadi MR, Pourmortazavib SM, Shandizc SAS, Ahmadide F, Batoolif H. 2014. Green synthesis of silver nanoparticles using Eucalyptus leucoxylon leaves extract and evaluating the antioxidant activities of extract. Nat Prod Res. 28:1964–1969.
- Patra JK, Dhal NK, Thatoi HN. 2011. In vitro bioactivity and phytochemical screening of Suaeda maritima (Dumort): a mangrove associate from Bhitarkanika, India. Asian Pac J Trop Med. 4:727–734.
- Prieto P, Nistora V, Nounehb K, Oyama M, Abd-Lefdil M, Diaz R. 2012. XPS study of silver, nickel and bimetallic silver–nickel nanoparticles prepared by seed-mediated growth. Appl Surf Sci. 258:8807–8813.
- Rajendran N, Subramaniam S, Raja MRC, Subbarao HMV, Raghunandan S, Venkatasubramanian U, et al. 2016. Design, synthesis and "in vitro" anti-leukemic evaluation of ferulic acid analogues as BCR-Abl inhibitors. RSC Adv. 6:70480–70484.
- Ravikumar S, Gnanadesigan M, Inbaneson JS, Kalaiarasi A. 2011. Hepatoprotective and antioxidant properties of Suaeda maritima(L.) Dumort ethanolic extract on concanavalin-A induced hepatotoxicity in rats. Indian J Exp Biol. 49:445–460.
- Saravanakumar A, Peng MM, Ganesh M, Jayaprakash J, Mohankumar M, Jang HT. 2016. Low-cost and eco-friendly green synthesis of silver nanoparticles using Prunus japonica (Rosaceae) leaf extract and their antibacterial, antioxidant properties. Artif Cells Nanomed Biotechnol. 1–7. doi: 10.1080/21691401.2016.1203795.
- Singh H, Du J, Yi TH. 2016. Biosynthesis of silver nanoparticles using Aeromonas sp. THG-FG1.2 and its antibacterial activity against pathogenic microbes. Artif Cells Nanomed Biotechnol. DOI: 10.3109/21691401.2016.1163715.
- Sinha SN, Paul D. 2015. Phytosynthesis of silver nanoparticles using Andrographis paniculata leaf extract and evaluation of their antibacterial activities. Spectrosc Lett. 48:600–604.
- Zhou JC, Wang X, Xue M, Xu Z, Hamasaki T, Yang Y, Wang K, Dunn B. 2010. Characterization of gold nanoparticle binding to microtubule filaments. Mater Sci Eng C. 30:20–26.