Abstract
miRNAs were reported as oncogene or tumour suppressors in various cancers and played important roles in tumour development and progression. Dysregulated miR-133 has been reported in several cancers, however, the expression and biological function of miR-133 in glioma remained unclear. In this study, we found that miR-133 expression level was significantly decreased in glioma tissues and cell lines by RT-qPCR. Then miR-133 mimics were used to evaluate the effects of miR-133 on cell proliferation and invasion in vitro. We found that overexpressed miR-133 could significantly suppress cell growth, and invasion in U87 cells. Additionally, we found that forkhead box C1 (FOXC1) was overexpressed in glioma tissue and it was directly regulated by miR-133. Overall, this study is the first proof to demonstrate that miR-133 function as tumour suppressor in glioma and inhibit cell proliferation and invasioned by directly targeting FOXC1, implying miR-133 as a potential therapeutic target for glioma.
Keywords:
Introduction
Human glioma, characterized by high metastasis ability, is the most common primary tumour of the central nervous system [Citation1–3]. Despite the improved therapy including surgery, radiation and chemotherapy, the survival rate of patients with high-grade glioma is less than 1 year [Citation4,Citation5]. Therefore, it is critical for investigating the molecular mechanism of glioma progression to develop novel and effective therapeutic strategies for glioma.
MicroRNAs (miRNAs) is a class of 17–27 nucleotides single-stranded small non-coding RNAs, and an accumulating studies showed that it can regulate gene expression by binding to its target of mRNAs at 3′ untranslated region (UTR) and inhibiting gene translation [Citation6,Citation7]. The miRNAs, as an intrinsic regulators, are known to involve in various cellular processes, including cell proliferation, differentiation, invasion and apoptosis [Citation8]. Accumulating studies revealed that miRNAs revealed, function as oncogenes or tumour suppressors, are dysregulated in various cancers and have been linked to the development and progression of various cancers, including breast cancer colorectal carcinoma, colorectal cancer, glioma and so on [Citation9–13]. Recently, dysregulated miRNAs was also reported in glioma [Citation14,Citation15]. For example, Chen et al. revealed that miR-24 function as tumour suppressor by regulating the proliferation and invasion of glioma [Citation16,Citation17]. Yang et al. showed that downregulated miR-136 promoted apoptosis of glioma cells induced by chemotherapy [Citation18]. Moreover, miR-661, microRNA-200b-3p, miR-24-3p, miR-27a-3p and microRNA-26b were also reported to play an important role in glioma progression [Citation19–22]. However, the expression and mechanism of miR-133 in glioma has remained to be elucidated.
In this study, we detected the expression levels of miR-133 in glioma tissues and cell lines. And then, we investigated the biological function and molecular mechanisms of miR-133 in regulating proliferation and invasion of glioma cells and elucidated the underlying mechanism, indicating miR-133 as novel diagnostic or therapeutic targets for glioma.
Materials and methods
Tissue samples
A total of 30 tumour and adjacent non-tumour tissues samples were collected from patients who had undergone surgical treatment in Shandong Provincial Hospital affiliated to Shandong University from 2013 to 2015. None of the patients had received chemotherapy or radiotherapy before surgery. This study was approved by the ethics committee of Shandong University, and the informed consent was obtained from each patient. Tissue fragments were immediately frozen in liquid nitrogen under ribonuclease-free conditions at the time of surgery and stored at −80 °C.
Cell culture and transfection
The glioma cell lines (U87) were gathered from the Cell Resource Center of Shanghai Institutes for Biological Sciences and cultured in DMEM (Sigma-Aldrich, Poole, Dorset, UK) supplemented with 10% fetal bovine serum (Biosera, Ringmer, UK), penicillin (100 IU/ml) and streptomycin (100 mg/ml; Sigma-Aldrich) in a humidified atmosphere at 37 °C with 5% CO2.The U87 cells were seeded in 6-well plates and then transfected with 50 nM of miR-133 mimics or inhibitors (GenePharma, Co., Ltd., Shanghai, China) using Lipofectamine® RNAiMAX (Invitrogen Life Technologies, Carlsbad, CA) as previously [Citation23].
Cell proliferation and invasion analysis
The U87 cells were seeded in 96-well microplates 24 h after transfection. Then, Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) was used to detect the ability to cell proliferation at 0, 24, 48 and 72 h according to the manufacturer’s instructions. The transwell invasion assay was used to detect cell migration capability as previous work [Citation24].
Western-blot analysis
Total protein was collected to use cell lysis reagent (Sigma-Aldrich, St. Louis, MO) and then separated by 10% SDS-PAGE and transferred into membranes (BD Biosciences, San Jose, CA), which were incubated with primary antibodies anti-GAPDH (ab8425), anti-RNF31 (ab46322) from Abcam (Cambridge, MA) at 4 °C overnight. Then incubated with goat anti-rabbit secondary antibody (Bioworld Technology, Inc., St. Louis Park, MN) under room temperature for 1 h. The protein signals were detected by ECL detection systems (SuperSignal West Femto, Pierce, Waltham, MA).
qRT-qPCR
We extracted RNA with Trizol solution (Sigma-Aldrich, St. Louis, MO) and then 1 μg RNA was used to reverse transcription and quantitative PCR which was performed as previously described [Citation25]. Primer sequences for quantitative PCR were used: miR-133 (sense) 5′-AGGCAAGATGCTGGCATAGCT-3′; RUNU6B was used as a control to normalize the expression level of miRNAs, and the primer (sense): 5′-GCGAGCACAGAATTAATACGAC-3′. FOXC1 forward, 5′-GGCGAGCAGAGCTACTACC-3′ and reverse, 5′-TGCGAGTACACGCTCATGG-3′; and GAPDH forward, 5′-TGTGGGCATCAATGGATTTGG-3′ and reverse, 5′-ACACCATGTATTCCGGGTCAAT-3′ was used as a control to normalize the expression level of mRNA.
Dual luciferase reporter assay
FOXC1–3′untranslated region (UTR) containing the putative miR-133 target sites was PCR amplified from the genomic DNA of a healthy control donor and cloned into the pGL3 Basic dual Luciferase reporter plasmid as described previously [Citation26]. Cells were co-transfected with either pGL3-WT-FOXC1–3′-UTR (wild type) or pGL3-Mut-FOXC1–3′-UTR (mutant) vector and the miR-133 mimic and then detected luciferase activities by Dual-Luciferase Reporter Assay system (Promega, Madison, WI) after 48 h.
Statistical analysis
Data are presented as the mean ± standard deviation. The differences were analyzed by using the Student's t-test. All the analyses were performed by using SPSS software, version 17.0 (SPSS Inc., Chicago, IL). p < .05 was considered to indicate a statistically significant difference.
Results
MiR-133 is downregulated in glioma tissues
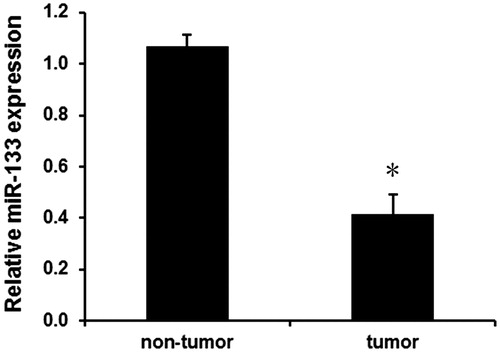
We first analyzed the expression levels of miR-133 in human glioma tissues and adjacent normal tissues using qPCR assay. As shown in , miR-133 expression was evidently downregulated in glioma tissues compared with the adjacent normal tissues (p < .01). Moreover, the relationship of overall survival with miR-133 expression was evaluated by Kaplan–Meier analysis and log–rank test. We found that the overall survival rate of patients with miR-133 low expression group was lower than those with miR-133 high expression level (). These findings suggest that downregulated miR-133 may involve in tumourigenesis of glioma.
MiR-133 expression and clinicopathologic features of glioma patients
The relationship between clinicopathological characteristics and miR-133 in glioma was analyzed, as shown in . We found that miR-133 expression was significantly associated with high grades III–IV, showing a little association with age, gender and tumour size instead of age, sex and tumour location.
Table 1. The relationship between miR-133 expression and clinicopathological features in glioma patients.
Effects of miR-133 on glioma cell proliferation and invasion
Based on these results, we speculated that miR-133 may regulate cell proliferation and invasion. Here, miR-133 mimics were used to transfect in glioma cells. As shown in , miR-133 mimic evidently increased miR-133 expression in U87 cells.
Figure 2. Effects of miR-133 on cell proliferation and invasion of glioma. (A) The effects of miR-133 mimic on miR-133 expression in U87 cells. The effects of miR-133 mimic on cell proliferation (B) and invasion (C and D) in U87 cells. *p < .05.
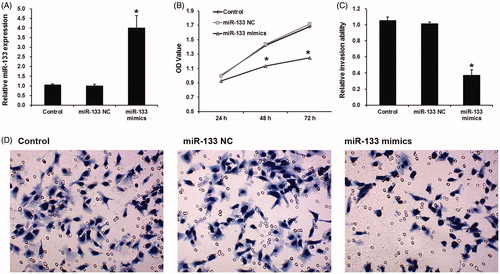
Next, we analyzed the effects of miR-133 mimics on glioma cell proliferation and invasion by using CCK-8 assay and wound healing assay. Results showed that miR-133 mimic led a significant reduction of cell proliferation () and migration () in U87 cells, indicating miR-133 may inhibit cell proliferation and migration in glioma.
FOXC1 was overexpressed in glioma tissue and directly regulated by miR-133
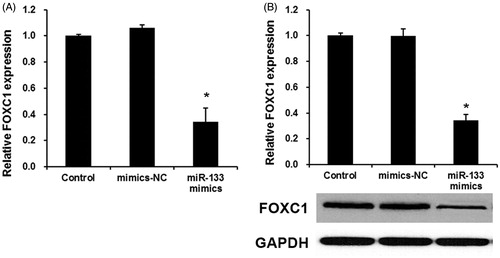
To reveal the mechanism of miR-133 in glioma progression, we used miRNA target prediction websites www.microRNA.org and found that miR-133 could bind to the 3′UTR of FOXC1 mRNA (). To further confirm this target role of miR-133, we used a dual-luciferase reporter assay was used. As shown in , miR-133 mimic significantly inhibited luciferase activity of the wild-type FOXC1–3′ UTR, but it failed to affect the luciferase activity of mutated target sites, suggesting that FOXC1 is a direct target gene of miR-133. Next, we analyzed the expression levels of FOXC1 in human PAs tissues and adjacent normal tissues by qPCR. As shown in , the expression levels of FOXC1 were significantly increased in glioma tissues compared with the adjacent normal tissues (p < .01), indicating that the upregulation of FOXC1 may be involved in human glioma carcinogenesis. Finally, the effect of miR-133 transfection on endogenous FOXC1 mRNA and protein expression was subsequently evaluated in U87 cells by qRT-PCR. As shown in , the expression levels of FOXC1 were markedly downregulated by miR-133 mimics. Collectively, our results demonstrated that miR-133 downregulated the expression of FOXC1 in glioma cells.
Figure 3. FOXC1 is a direct target of miR-133 in glioma. (A) miRNA target soft predicted the target site of miR-133 on the 3’UTR of FOXC1 mRNA. (B) dual-luciferase reporter assay revealed the target role of miR-133 on FOXC1.
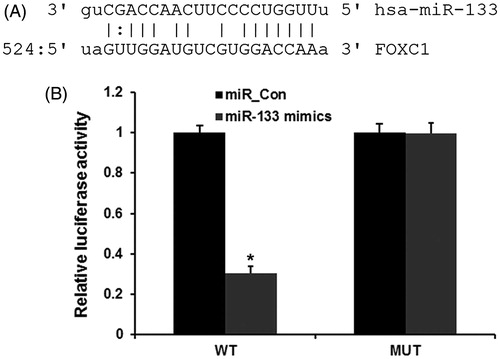
Figure 4. FOXC1 was overexpressed in glioma tissue and directly regulated by miR-133. (A) qRT-PCR assay the FOXC1 expression in glioma tissues. *p < .01 versus normal tissues. (B) The effects of miR-133 mimics on the expression levels of FOXC1 mRNA (B) and protein (C). *p < .01 versus control group.
Discussion
In this study, the roles of miR-133 in glioma progression were detected. In this present study, we firstly detected the expression levels of miR-133 in glioma and found that miR-133 expression was evidently downregulated in glioma tissues comparing with the adjacent normal tissues. Moreover, clinicopathologic features of glioma patients revealed that miR-133 expression was significantly associated with high grades III–IV. Finally, we analyzed its effect on proliferation and invasion of glioma. Advanced in vitro study demonstrated that miR-133 produced a capital effect through inhibiting the expression of FOXC1.
Accumulating studies showed that the dysregulated miRNAs, as a function of oncogenes and tumour suppressors, were involved in the development and progression of various human cancers, including glioma [Citation27]. Li et al. found that overexpression of miR-661 obviously suppressed the proliferation, migration and invasion of glioma cells [Citation19]. Wang et al. also found that miR-30a-5p promotes glioma cell growth invasion by repressing NCAM [Citation15]. MiR-200b-3p, which was found to be downregulated in glioma tissues and human glioma cells, was found to suppress glioma tumour growth, invasion and reverses EMT [Citation13]. Additionally, downregulated miR-136 was absorved in human glioma, and that the miRNA promoted apoptosis of glioma cells induced by chemotherapy [Citation18,Citation28], miR24-3p and miR27a-3p function as oncogene and promote cell proliferation [Citation21]. In total, the previous reports indicated miRNAs as potential bio-markers in glioma. miR-133 was reported in dysregulated in several kinds of cancers. For example, miR-133 was downregulated in gastric cancer tissues, which was negatively associated with tumour size, invasion depth and peripheral organ metastasis [Citation29,Citation30]. miR-133a also was downregulated in cervical cancer tissues and cell lines, and the aberrant expression of miR-133a was correlated with lymph node metastasis, histological grade [Citation31]. miR-133 inhibited cell proliferation, migration and invasion of prostate cancer and bladder cancer [Citation32,Citation33]. In this study, our data revealed that miR-133 greatly inhibited cellular proliferation of glioma cancer cells. Furthermore, up-regulation of miR-133 could inhibit U87 cancer cell invasion and it suggested that miR-133 may contribute to metastasis in laryngeal cancer. The anti-cancer effect of miR-133 might from its ability of inhibiting the expression of FOXC1.
FOXC1 is a member of Forkhead box family transcription factors, which is defined by a common DNA-binding domain (DBD) termed the forkhead box or winged helix domain [Citation34]. Recently, FOXC1 was reported to be dysregulated in various cancers and played an important role on tumour progression. For example, FOXC1 was described as an important prognostic biomarker in basal-like breast cancer [Citation35], gastric cancer [Citation36], non-small cell lung cancer [Citation37]. And FOXC1 was reported to involve migration and invasion of endometrial cancer [Citation38] and pancreatic cancer [Citation39]. However, the expression and mechanism of FOXC1 in glioma remain unclear. In this study, target prediction and dual-luciferase reporter assay revealed that FLOT2 was a direct target of miR-133 in glioma. Moreover, FLOT2 was upregulated in glioma tissue and the expressed levels of FOXC1 were markedly downregulated by miR-133 mimics. Taken together, these findings support a novel role for miR-133 in invasion and proliferation of glioma cells. Thus, miR-29b may be used as a new potential therapeutic target for glioma. However, only in vitro studies were conducted in this study, we would conduct several more studies to detect the influence of miR-133 of FOXC1 expression in vivo. In summary, we firstly investigated the role of miR-133 in glioma and revealed miR-133 as a tumour suppressor which repressed glioma cell proliferation and invasion by targeting FOXC1, indicating a novel therapeutic target for the treatment of glioma.
Disclosure statement
The authors declare no conflict of interest.
References
- Wu X, Hu A, Zhang M, et al. Effects of Rab27a on proliferation, invasion, and anti-apoptosis in human glioma cell. Tumor Biol. 2013;34:2195–2203.
- Xu Y, Xu W, Lu T, et al. miR-126 affects the invasion and migration of glioma cells through GATA4. Artif Cells Nanomed Biotechnol. Forthcoming. [7 p.]. doi: 10.1080/21691401.2016.1226179
- Liu Q, Liao F, Wu H, et al. Different expression of miR-29b and VEGFA in glioma. Artif Cells Nanomed Biotechnol. 2016;44:1927–1932.
- Nieder C, Adam M, Molls M, et al. Therapeutic options for recurrent high-grade glioma in adult patients: recent advances. Crit Rev Oncol Hematol. 2006;60:181–193.
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996.
- Dou Y, Wang Y, Xu J, et al. Experiment research on inhibition of glioma with sTRAIL in vitro. Artif Cells Nanomed Biotechnol. 2014;42:186–191.
- Jinling W, Sijing S, Jie Z, et al. Prognostic value of circulating microRNA-21 for breast cancer: a systematic review and meta-analysis. Artif Cells Nanomed Biotechnol. Forthcoming. [6 p.]. doi: 10.1080/21691401.2016.1216856
- Xu XM, Qian JC, Deng ZL, et al. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett. 2012;4:339–345.
- Chen S, Dai Y, Zhang X, et al. Increased miR-449a expression in colorectal carcinoma tissues is inversely correlated with serum carcinoembryonic antigen. Oncol Lett. 2014;7:568–572.
- Lu Z, Ye Y, Jiao D, et al. miR-155 and miR-31 are differentially expressed in breast cancer patients and are correlated with the estrogen receptor and progesterone receptor status. Oncol Lett. 2012;4:1027–1032.
- Lei SL, Zhao H, Yao HL, et al. Regulatory roles of microRNA-708 and microRNA-31 in proliferation, apoptosis and invasion of colorectal cancer cells. Oncol Lett. 2014;8:1768–1774.
- Luo J, Zhou J, Cheng Q, et al. Role of microRNA-133a in epithelial ovarian cancer pathogenesis and progression. Oncol Lett. 2014;7:1043–1048.
- Wu J, Cui H, Zhu Z, et al. MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and inhibits tumor growth of glioma through down-regulation of ERK5. Biochem Biophys Res Commun. 2016;478:1158–1164.
- Wei J, Wang F, Kong LY, et al. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73:3913–3926.
- Wang Z, Dai X, Chen Y, et al. miR-30a-5p is induced by Wnt/β-catenin pathway and promotes glioma cell invasion by repressing NCAM. Biochem Biophys Res Commun. 2015;465:374–380.
- Cai JJ, Qi ZX, Chen LC, et al. miR-124 suppresses the migration and invasion of glioma cells in vitro via Capn4. Oncol Rep. 2016;35:284–290.
- An L, Liu Y, Wu A, et al. microRNA-124 inhibits migration and invasion by down-regulating ROCK1 in glioma. PLoS One. 2013;8:e69478.
- Yang Y, Wu J, Guan H, et al. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. FEBS Lett. 2012;586:3608–3612.
- Li Z, Liu YH, Diao HY, et al. MiR-661 inhibits glioma cell proliferation, migration and invasion by targeting hTERT. Biochem Biophys Res Commun. 2015;468:870–876.
- Wu J, Cui H, Zhu Z, et al. MicroRNA-200b-3p suppresses epithelial–mesenchymal transition and inhibits tumor growth of glioma through down-regulation of ERK5. Biochem Biophys Res Commun. 2016;478:1158–1164.
- Xu W, Liu M, Peng X, et al. miR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Int J Oncol. 2013;42:757–766.
- Wu N, Zhao X, Liu M, et al. Role of microRNA-26b in glioma development and its mediated regulation on EphA2. PLoS One. 2011;6:e16264.
- Ehrlund A, Anthonisen EH, Gustafsson N, et al. E3 ubiquitin ligase RNF31 cooperates with DAX-1 in transcriptional repression of steroidogenesis. Mol Cell Biol. 2009;29:2230–2242.
- Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333.
- Zhu J, Zhao C, Zhuang T, et al. RING finger protein 31 promotes p53 degradation in breast cancer cells. Oncogene. 2016;35:1955–1964.
- Micale L, Fusco C, Fontana A, et al. TRIM8 downregulation in glioma affects cell proliferation and it is associated with patients survival. BMC Cancer. 2015;15:470.
- Fakhoury M. Drug delivery approaches for the treatment of glioblastoma multiforme. Artif Cells Nanomed Biotechnol. 2016;44:1365–1373.
- Chen W, Yang Y, Chen B, et al. MiR-136 targets E2F1 to reverse cisplatin chemosensitivity in glioma cells. J Neurooncol. 2014;120:43–53.
- Cheng Z, Liu F, Wang G, et al. miR-133 is a key negative regulator of CDC42-PAK pathway in gastric cancer. Cell Signal. 2014;26:2667–2673.
- Zhang XT, Zhang Z, Xin YN, et al. Impairment of growth of gastric carcinoma by miR-133-mediated Her-2 inhibition. Tumor Biol. 2015;36:8925–8930.
- Song X, Shi B, Huang K, et al. miR-133a inhibits cervical cancer growth by targeting EGFR. Oncol Rep. 2015;34:1573–1580.
- Zhou Y, Wu D, Tao J, et al. MicroRNA-133 inhibits cell proliferation, migration and invasion by targeting epidermal growth factor receptor and its downstream effector proteins in bladder cancer. Scand J Urol. 2013;47:423–432.
- Tao J, Wu D, Xu B, et al. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967–1975.
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859.
- Jin Y, Han B, Chen J, et al. FOXC1 is a critical mediator of EGFR function in human basal-like breast cancer. Ann Surg Oncol. 2014;21:S758–S766.
- Xu Y, Shao QS, Yao HB, et al. Overexpression of FOXC1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2014;64:963–970.
- Wei LX, Zhou RS, Xu HF, et al. High expression of FOXC1 is associated with poor clinical outcome in non-small cell lung cancer patients. Tumor Biol. 2013;34:941–946.
- Chung TK, Lau TS, Cheung TH, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130:1036–1045.
- Yu C, Wang M, Li Z, et al. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell Oncol (Dordr). 2015;38:173–181.