Abstract
Non-coding RNAs play an important role in the pathogenesis of prostate cancer (PC). This study aims to characterize the role of GAS5 rs145204276 and HOTAIR rs4759314 polymorphisms in the pathogenesis of PC. Both INS allele of GAS5 rs145204276 and A allele of HOTAIR rs4759314 were identified to increase the survival of PC patients. And patients carrying DEL/DEL + AG genotypes tend to present higher levels of HMGB1, GAS5, HOTAIR and lower levels of miR-1284 and miR-22. In addition, the transcription activity of GAS5 promoter was increased by the deletion allele of rs145204276 polymorphism, while the G allele of rs4759314 polymorphism increased the transcription activity of HOTAIR promoter. GAS5 and HOTAIR could bind to miR-1284 and miR-22, respectively, while miR-1284 and miR-22 could bind to the 3′UTR of HMGB1. Compared with the control group, the expressions of miR-1284 or miR-22 were decreased with the presence of GAS5 or HOTAIR, and the expression of HMGB1 was the highest in the GAS5 + HOTAIR group. In summary, the findings of this study demonstrated that both GAS5 rs145204276 and HOTAIR rs4759314 polymorphisms could affect the prognosis of PC by modulating the expression of HMGB1 via modulating the GAS5/miR-1284/HMGB1 and HOTAIR/miR-22/HMGB1 signalling pathways.
Introduction
As previous reported, prostate cancer (PC) is the second ranked malignancy and the fifth major cause of mortality worldwide, which is attributed to almost 20% of cancers in male patients [Citation1]. Histopathological assessment of tumour tissue samples was usually utilized to confirm the diagnosis of PC. However, biopsy is associated with certain disadvantages such as infection and bleeding [Citation2].
Enriched in chromatin as a non-histone component, high mobility group box 1 (HMGB1) is released from cells into the extracellular environment, where HMGB1 binds to the receptor for advanced glycation end products (RAGE) and subsequently activates several critical cell signalling pathways, including p44/42MAPKs, p38 and NF-κB signalling, to promote cancer development and metastasis [Citation3–9]. In addition, upregulated HMGB1 expression was frequently observed in PC cases, although the value of HMGB1 in the prognosis of PC was only studied in one article, which showed that the expression of HMGB1 protein acted as a prognostic factor for post-radical prostatectomy (RP) survival [Citation9–12]. Furthermore, it is also confirmed that the abnormality in HMGB1 and RAGE expression was involved in the progression of PC. As a result, the co-expression of HMGB1 and RAGE could be used as a better prognostic factor than each protein alone for the progression of PC.
MicroRNAs (miRNAs), which are short ncRNAs of about 22 nucleotides in length, and long noncoding RNAs (lncRNAs), which are usually more than 200 nucleotides in length, can simultaneously regulate the expression of their target genes by modulating the translation and transcription of their target mRNAs. On the other hand, mRNAs also affect the expression of ncRNAs in specific ways [Citation13,Citation14]. For example, the HOX transcript antisense RNA (HOTAIR), which is located on human chromosome 12, encodes a lncRNA of about 2 kb that is enriched in multiple cancers, including laryngeal, pancreas, liver, colorectal and breast cancer [Citation15,Citation16]. In addition, HOTAIR expression is upregulated in PC cell lines resistant to castration, whereas the silence of HOTAIR expression reduced the viability of PC cells, suggesting that HOTAIR plays an essential role in the growth of PC cells [Citation16].
In past studies, genetic variants in the promoter of lncRNAs were shown to regulate the expression of their host lncRNAs through methylation [Citation17,Citation18]. For example, as a 5 bp indel polymorphism (-/AGGCA) located in the promoter of GAS5, rs145204276 was reported to increase the risk of hepatocellular carcinoma (HCC) by regulating GAS5 expression [Citation18]. The potential role of rs145204276 in the prognosis of GC has also been reported. For example, patients carrying the del allele of rs145204276 have significantly better survival and lower rates of cancer progression and metastasis. Therefore, it is likely that rs145204276 plays a protective role in the pathogenesis of GC by regulating GAS5 expression. Furthermore, it has been shown that the G allele of the rs4759314 polymorphism located in HOTAIR promoter significantly increased the risk of CHD by decreasing the transcription efficiency of HOTAIR promoter and subsequently reducing the expression of HOTAIR.
HMGB1 has been implicated in the pathogenesis and prognosis of PC [Citation19,Citation20]. Meanwhile, it was reported that both miR-1284 and miR-22 function as regulators of HMGB1 expression [Citation21,Citation22]. In addition, GAS5 and HOTAIR play a role as competing endogenous RNAs against miR-1284 and miR-22, respectively [Citation21,Citation23]. Furthermore, the rs145204276 and rs4759314 polymorphisms located in the promoters of GAS5 and HOTAIR, respectively, can affect the transcription efficiency of these promoters [Citation24,Citation25]. Therefore, the two polymorphisms may affect the prognosis of PC via modulating the GAS5/miR-1284/HMGB1 and HOTAIR/miR-22/HMGB1 signalling pathways. In this study, we collected samples from PC patients to probe the effect of above two polymorphisms on the prognosis of PC.
Materials and methods
Patients and sample collection
A total of 159 PC patients participated in this study. Clinicopathological information of all subjects, including their age, PSA level, Gleason score, T stage, N stage, and the presence of distant metastasis, were collected and compared, as listed in . Tissue and peripheral blood samples were collected from all patients, and the patients were then divided into four groups according to their genotypes of rs145204276 polymorphism of GAS5 (INS/INS, INS/DEL and DEL/DEL) and rs4759314 polymorphism of HOTAIR (AA/AG): a GAS5 rs145204276 DEL/DEL + HOTAIR rs4759314 AG group (Group 1, N = 26); a GAS5 rs145204276 DEL/DEL + HOTAIR rs4759314 AA group (Group 2, N = 36); a GAS5 rs145204276 INS/INS and INS/DEL + HOTAIR rs4759314 AG group (Group 3, N = 42); and a GAS5 rs145204276 INS/INS and INS/DEL + HOTAIR rs4759314 AA group (Group 4, N = 55). This study was approved by the ethics committee of our institute, and all patients have signed informed consent prior to the start of this study.
Table 1. Characteristics of patients diagnosed with prostate cancer recruited in this present study.
Taqman genotyping
The genotypes of rs145204276 polymorphism of GAS5 (INS/INS, INS/DEL and DEL/DEL) and rs4759314 polymorphism of HOTAIR in collected tissue and peripheral blood samples were determined using a Taqman genotyping assay (ABI, Foster City, CA) following the instruction of the manufacturer. The assay was performed on a Roche LightCycler 480 II PCR machine (Roche, Mannheim, Germany) and the results were analysed by Version 1.5 of LightCycler 480 Software (Roche, Mannheim, Germany). Each experiment was repeated three times.
RNA isolation and real-time PCR
A miRNeasy Mini kit (Qiagen, Valencia, CA) was used to isolate total RNA from collected tissue and peripheral blood samples following the instruction of the kit. The concentration and quality of isolated RNA were quantified using a Nano Drop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Subsequently, 10 μg total RNA of each sample were reversely transcribed into cDNA using a reverse transcription kit (Invitrogen, Carlsbad, CA). The conditions of reverse transcription reactions were: 25 °C for 30 min, 42 °C for 40 min and 85 °C for 5 min. In the next step, the expression of HMGB1 mRNA, GAS5, miR-1284, HOTAIR and miR-22 was measured using real-time PCR with specifically designed primers. The conditions of RT-PCR reactions were 94 °C for 10 min, followed by 45 cycles of 94 °C for 15 s and 60 °C for 60 s. Finally, the relative expression levels of HMGB1 mRNA (Forward: 5′- GCGAAGAAACTGGGAGAGATGTG-3′; Reverse: 5′- GCATCAGGCTTTCCTTTAGCTCG-3′), GAS5 (Forward: 5′- CCCAAGGAAGGATGAG-3′; Reverse: 5′-ACCAGGAGCAGAACCA-3′), miR-1284 (Forward: 5′- TACAGACCCTGGCTTTTC-3′; Reverse: 5′- GAACATGTCTGCGTATCTC-3′), HOTAIR (Forward: 5′- CCAGAGAACGCTGGAAAAACCTG-3′; Reverse: 5′- GGAGATGATAAGAAGAGCAAGGAA-3′), and miR-22 (Forward: 5′- GTTCTTCAGTGGCAAGC-3′; Reverse: 5′-GAACATGTCTGCGTATCTC-3′) were quantified using the 2−ΔΔCT method and GAPDH (Forward: 5′- GTCTCCTCTGACTTCAACAGCG-3′; Reverse: 5′- ACCACCCTGTTGCTGTAGCCAA -3′) expression as the internal control. All RT-PCR reactions were run in triplicate and each experiment was repeated at least three times.
Cell culture and transfection
LNCaP and PC3 cells were cultured in a Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% foetal bovine serum (FBS) (Invitrogen, Thermo Fisher Scientific, Waltham, MA), 4.5 g/L of glucose (Sigma-Aldrich, St. Louis, MO), 1.5 g/L of sodium bicarbonate (Sigma-Aldrich, St. Louis, MO), 4 mM of L-glutamine (Gibco, Thermo Fisher Scientific, Waltham, MA), 65 units/mL of streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA) and 100 units/mL penicillin (Gibco, Thermo Fisher Scientific, Waltham, MA). The cells were maintained at 37 °C and saturated humidity in an atmosphere containing 5% CO2 and 95% air. Subsequently, LNCaP and PC3 cells were seeded into 24-well plates at a density of 3 × 105 cells/well and cultured overnight before transfection. On the following day, the cells were divided into four groups, i.e. a NC group (cells transfected with a negative control plasmid), a pGL3-GAS5 group (cells transfected with a pGL3-GAS5 plasmid), a pGL3-HOTAIR group (cells transfected with a pGL3-HOTAIR plasmid) and a pGL3-GAS5 + pGL3-HOTAIR group (cells co-transfected with both pGL3-GAS5 and pGL3-HOTAIR plasmids). The transfection was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the instruction of the manufacturer. At 48-h post-transfection, all transfected cells were harvested and subjected to subsequent real-time PCR or Western blot analyses. Each experiment was carried out in triplicate and repeated for at least three times.
Vector construction, mutagenesis and luciferase assay
PCR was performed to amplify the sequences of the promoters of GAS5 and HOTAIR, respectively, and the PCR products were inserted to separate pGL3 firefly luciferase reporter vectors (Promega, Madison, WI). At the same time, a quick-change site-directed mutagenesis Kit (Stratagene, La Jolla, CA) was used to introduce mutations into the rs145204276 and rs4759314 polymorphisms located on the promoters of GAS5 and HOTAIR, respectively, so as to create mutant vectors of GAS5 and HOTAIR, respectively. In the next step, in order to probe the effect of rs145204276 and rs4759314 polymorphisms on the transcription efficiency of GAS5 and HOTAIR promoters, LNCaP and PC3 cells were seeded into 24-well plates at a density of 3 × 105 cells/well and then transfected with either wild type or mutant vectors of GAS5 and HOTAIR, respectively, for 48 h. At 48 h post-transfection, the luciferase activity of transfected cells was measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the instruction of the kit. Similarly, in order to assay the regulatory relationships of GAS5 versus miR-1284, HOTAIR versus miR-22, HMGB1 versus miR-1284 and HMGB1 versus miR-22, the Quick-Change Site-Directed Mutagenesis Kit was used to introduce mutations in GAS5 and HMGB1 containing the binding sites of miR-1284 and miR-22, respectively, as well as in the sequence of miR-22 containing the binding site of HOTAIR, to generate mutant pGL3 vectors for GAS5, miR-22, and HMGB1, respectively. In the next step, LNCaP and PC3 cells were co-transfected with wild type/mutant vectors of GAS5/miR-1284/HMGB1 and HOTAIR/miR-22/HMGB1, respectively, and the luciferase activity of transfected cells was measured at 48 h post-transfection using the Dual-Luciferase Reporter Assay System. Each experiment described above was carried out in triplicate and repeated for at least three times.
Western blot analysis
To measure the protein expression of HMGB1, cell and tissue samples were lysed in a lysis buffer before a Bradford reagent (Bio-Rad Laboratories, Hercules, CA) was used to determine the concentration of isolated protein following a standard protocol. SDS-PAGE was then used to resolve sample proteins, which were subsequently transferred onto nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ) and blocked using Tris-buffered saline containing 5% non-fat dry milk and 0.1% Tween20. In the next step, the membrane was incubated at 4 °C for 12 h with primary antibodies against HMGB1 (1:1000 dilution, Abcam, Cambridge, MA) and β-actin (internal control, 1:10,000 dilution, Abcam, Cambridge, MA), followed by incubation with horseradish peroxidase (HPR)-labelled secondary antibodies (1:10,000 dilution, Abcam, Cambridge, MA) at room temperature for 1 h. Subsequently, the membrane was visualized using an enhanced chemiluminescence kit (Thermo Fisher Scientific, Waltham, MA) and the relative expression of HMGB1 protein was calculated using β-actin as the internal control. Each experiment was repeated three times.
Immunohistochemistry
Tissue samples were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin and sectioned into 4 μm slices. Subsequently, the sections were incubated in 3% H2O2 for 10 min to block endogenous peroxidase and then incubated with anti-MMP-2 (1:500 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-HMGB1 primary antibodies (1:1000 dilution, Abcam, Cambridge, MA) for 12 h at 4 °C. Subsequently, horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2000 dilution, GE Healthcare, Logan, UT) were used to incubate the sections for 2 h at room temperature. A DAB (3, 3-diaminobenzidine) substrate kit (Vector Laboratories Inc., Burlingame, CA) was then used to stain the samples in accordance with the guideline of the supplier, followed by counterstaining with haematoxylin and observation underneath an inverted microscope. All experiments were repeated at least three times.
Statistical analysis
All data were shown as mean ± SEM. Student t-tests were used to evaluate the statistical differences between two groups, while the comparisons among multiple groups were performed using one-way ANOVA (Turkey’s test as post hoc test). Furthermore, logistic regression analysis was used to analyse the association between the genotypes of polymorphisms and the risk of PC. ROC curves of survival were plotted and analysed by Cox regression and Kaplan–Meier analyses using NCSS statistical software (Kaysville, UT). All statistical analyses were performed using SPSS 21.0 (IBM, Chicago, IL), and a p value of <.05 was considered to be statistically significant.
Results
Clinicopathological characteristics of patients
Student t tests were performed to compare the patient groups and the results revealed no obvious difference among these four groups in terms of above clinicopathological characteristics ().
Genotypic association between rs145204276/rs4759314 and the survival of PC
Cox regression and Kaplan–Meier analyses were used to assess whether GAS5 rs145204276 and HOTAIR rs4759314 polymorphisms played an important role in the survival of PC. As shown in , the overall survival of PC was the lowest in Group 1 and the highest in Group 4, indicating that the INS genotype of rs145204276 polymorphism in GAS5 and the A allele of rs4759314 polymorphism in HOTAIR apparently increased the survival rate of PC patients.
Figure 1. Genotypic association between rs145204276/rs4759314 and the survival of PC (Group A: GAS5 rs145204276 DEL/DEL + HOTAIR rs4759314 AG; Group B: GAS5 rs145204276 DEL/DEL + HOTAIR rs4759314 AA; Group C: GAS5 rs145204276 INS/INS&INS/DEL + HOTAIR rs4759314 AG; Group D: GAS5 rs145204276 INS/INS&INS/DEL + HOTAIR rs4759314 AA). (A) The INS genotype of rs145204276 polymorphism in GAS5 and the A allele of rs4759314 polymorphism in HOTAIR apparently increased the survival rate of PC patients; (B) The mRNA level of HMGB1 was the highest in Group 1 and the lowest in Group 4; (C) The level of HMGB1 protein was the highest in Group 1 and the lowest in Group 4; (D) The level of GAS5 mRNA in Groups 1 and 2 was much higher than that in Groups 3 and 4; (E) The level of miR-1284 in Groups 1 and 2 was apparently lower than that in Group 3 and Group 4; (F) The level of HOTAIR mRNA in Groups 1 and 3 was much higher than that in Group 2 and Group 4; (G) The level of miR-22 in Groups 1 and 3 was apparently lower than that in Group 2 and Group 4.
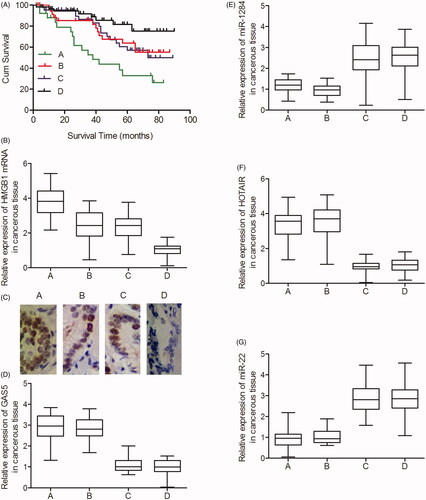
Expressions of GAS5, HOTAIR, miR-1284, miR-22 and HMGB1 among different genotypes
Real-time PCR and IHC assays were performed to investigate the effect of GAS5 rs145204276 and HOTAIR rs4759314 polymorphisms on the expression of GAS5, HOTAIR, miR-1284, miR-22 and HMGB1 in patients from the above four groups. As shown in , the levels of HMGB1 mRNA () and protein () were the lowest in Group 4 and the highest in Group 1. In addition, the levels of GAS5 () were higher in Group 1 and Group 2 than those in Groups 3 and 4. On the other hand, the levels of miR-1284 () were lower in Groups 1 and 2 than those in Groups 3 and 4. Furthermore, Groups 1 and 3 showed higher levels of HOTAIR () and lower levels of miR-22 () than Groups 2 and 4.
Interactions among GAS5, HOTAIR, miR-1284, miR-22 and HMGB1
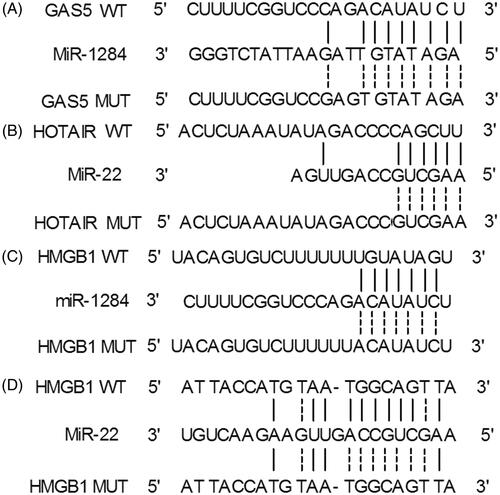
By searching several databases commonly used for target gene prediction (http://targetscan.org/), binding sites for GAS5 and HOTAIR were identified on miR-1284 () and miR-22 (), respectively. Meanwhile, the 3′UTR of HMGB1 contained binding sites for both miR-1284 () and miR-22 ().
Figure 2. Schematic comparison between GAS5 and miR-1284, HOTAIR and miR-22, and miR-1284/miR-22 and HMGB1. (A) A putative binding site of GAS5 was located in miR-1284; (B) A putative binding site of HOTAIR was located in miR-22; (C) Schematic representation of the miR-1284 binding site located in the 3′UTR of HMGB1; (D) Schematic comparison between miR-22 sequence and the wild-type/mutant “seed sequence” of miR-22 located in the 3′UTR of HMGB1.
We utilized luciferase assays to examine the effects of GAS5 rs145204276 and HOTAIR rs4759314 polymorphisms on the promoter activity of GAS5 and HOTAIR, respectively. In comparison with the transfection of control constructs, the transfection with the GAS5 constructs carrying the insert or deletion allele of rs145204276 polymorphism apparently increased the luciferase activity of LNCaP () and PC3 () cells. Moreover, compared with the insert allele of rs145204276 polymorphism, the presence of the deletion allele significantly promoted the transcription of GAS5 promoter. Similarly, in comparison with the transfection of control constructs, the transfection with HOTAIR constructs carrying the A or G allele of rs4759314 polymorphism apparently increased the luciferase activity of LNCaP () and PC3 () cells. Moreover, compared with the A allele of rs4759314 polymorphism, the presence of the G allele significantly promoted the transcription of HOTAIR promoter. In addition, the transfection with miR-1284 mimics reduced the luciferase activity of wild-type GAS5 but not that of mutant GAS5 in LNCaP () and PC3 () cells, while the transfection with miR-22 mimics reduced the luciferase activity of wild-type HOTAIR but not that of mutant HOTAIR in LNCaP () and PC3 () cells. Finally, the transfection with miR-1284 ( and ) and miR-22 ( and ) mimics reduced the luciferase activity of wild-type HMGB1 3′UTR but not that of mutant HMGB1 3′UTR in LNCaP () and PC3 () cells.
Figure 3. Interactions among GAS5, HOTAIR, miR-1284, miR-22 and HMGB1 in LNCaP cells. (A) Transfection with the insert or deletion allele of rs145204276 polymorphism increased the luciferase activity of GAS5, and the effect of rs145204276 deletion allele was much stronger than that of rs145204276 insert allele; (B) Luciferase activity of the cells was apparently increased after the transfection with A or G allele of HOTAIR rs4759314 polymorphism, and the presence of G allele of rs4759314 polymorphism more significantly promoted the transcription of HOTAIR promoter than the A allele; (C) Luciferase activity of wild-type GAS5 was decreased by transfecting the cells with miR-1284 mimics; (D) Luciferase activity of wild-type HOTAIR was down-regulated by transfecting the cells with miR-1284 mimics; (E) Luciferase activity of wild-type 3′UTR of HMGB1 but not that of mutant 3′UTR of HMGB1 was reduced by transfecting the cells with miR-1284 mimics; (F) Luciferase activity of wild-type 3′UTR of HMGB1 but not that of mutant 3′UTR of HMGB1 was reduced by transfecting the cells with miR-22 mimics; (G) The level of miR-1284 was decreased following the transfection with GAS5 or GAS5 + HOTAIR; (H) The level of miR-22 was reduced in cells transfected with HOTAIR or GAS5 + HOTAIR; (I) The level of HMGB1 mRNA was the highest in the GAS5 + HOTAIR group and the lowest in the control group; (J) The level of HMGB1 protein was the highest in the GAS5 + HOTAIR group and the lowest in the control group.
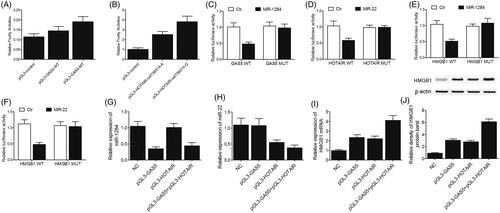
Figure 4. Interactions among GAS5, HOTAIR, miR-1284, miR-22 and HMGB1 in PC3 cells. (A) Transfection with the insert or deletion allele of rs145204276 polymorphism increased the luciferase activity of GAS5, and the effect of rs145204276 deletion allele was much stronger than that of rs145204276 insert allele; (B) Luciferase activity of the cells was apparently increased after the transfection with A or G allele of HOTAIR rs4759314 polymorphism, and the presence of G allele of rs4759314 polymorphism more significantly promoted the transcription of HOTAIR promoter than the A allele; (C) Luciferase activity of wild-type GAS5 was decreased by transfecting the cells with miR-1284 mimics; (D) Luciferase activity of wild-type HOTAIR was down-regulated by transfecting the cells with miR-1284 mimics; (E) Luciferase activity of wild-type 3′UTR of HMGB1 but not that of mutant 3′UTR of HMGB1 was reduced by transfecting the cells with miR-1284 mimics; (F) Luciferase activity of wild-type 3′UTR of HMGB1 but not that of mutant 3′UTR of HMGB1 was reduced by transfecting the cells with miR-22 mimics; (G) The level of miR-1284 was decreased following the transfection with GAS5 or GAS5 + HOTAIR; (H) The level of miR-22 was reduced in cells transfected with HOTAIR or GAS5 + HOTAIR; (I) The level of HMGB1 mRNA was the highest in the GAS5 + HOTAIR group and the lowest in the control group; (J) The level of HMGB1 protein was the highest in the GAS5 + HOTAIR group and the lowest in the control group.
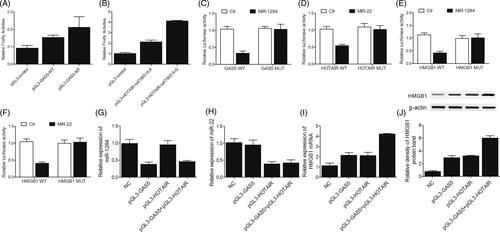
Real-time PCR and Western-blot analysis were then carried out to further confirm the interactions among GAS5, HOTAIR, miR-1284, miR-22 and HMGB1 in LNCaP () and PC3 () cells transfected with constructs carrying GAS5, HOTAIR or GAS5 + HOTAIR. Compared with the transfection of control vectors, the transfection of GAS5 and GAS5 + HOTAIR decreased the level of miR-1284 in LNCaP () and PC3 () cells. Similarly, compared with the transfection of control vectors, the transfection of HOTAIR and GAS5 + HOTAIR decreased the level of miR-22 in LNCaP () and PC3 () cells. Finally, the transfection with GAS5, HOTAIR and GAS5 + HOTAIR significantly increased the mRNA ( and ) and protein ( and ) levels of HMGB1 in LNCaP () and PC3 () cells, and GAS5 + HOTAIR exerted a stronger effect than GAS5 or HOTAIR alone on the expression of HMGB1.
Discussion
In this study, we enrolled 159 PC patients and found that the INS genotype of GAS5 rs145204276 polymorphism and the A allele of HOTAIR rs4759314 polymorphism both apparently increased the survival rate of PC patients.
Growing evidence has shown that HOTAIR can regulate key signalling pathways involved in cancer metastasis and invasion. For example, HOTAIR was shown to increase the metastasis and invasiveness of breast cancer by enhancing the expression of LAMC2, LAMB3 and ABL2 SNAIL, all of which were reported to enhance cancer metastasis [Citation15]. On the other hand, upregulated expression of HOTAIR in HCC may be used as a prognostic factor for post-hepatectomy recurrence of HCC by regulating the expression of VEGF and MMP9 [Citation26]. The above results suggest that HOTAIR plays an essential role in the progression of PC. Moreover, the rs4759314 polymorphism located in the promoter of HOXC11can affect the expression of HOXC11 by regulating its transcription efficiency [Citation26]. In fact, compared with the A allele of the rs4759314 polymorphism, the G allele was shown to increase the expression of HOTAIR, suggesting that the G allele is associated with a higher risk of carcinogenesis.
It was shown that GAS5 lncRNA can regulate both the apoptosis and proliferation of PC cells when these cells are exposed to a wide range of apoptotic stimuli. Therefore, GAS5 may be closely related to the carcinogenesis of PC and hence may be exploited as a target gene in PC therapy [Citation27]. It was reported that an increased level of GAS5 expression reduced the survival of PC cells in a manner similar to exposing the cells to 22Rv1. Located in the promoter region of GAS5, rs145204276 was associated with the altered expression of GAS5 in vitro [Citation28]. In previous studies, the DEL allele of rs145204276 was shown to increase the expression of GAS in both HCC and colorectal cancer [Citation28,Citation29]. Tao et al. [Citation28] hypothesized that the rs145204276 polymorphism can affect the transcriptional activity of GAS5 and hence regulate its expression by methylating CpG islands located in the GAS5 promoter. Furthermore, it was also found that gastric cancer patients carrying the del allele of rs145204276 were associated with a remarkably higher level of GAS5 expression in the tumour tissue. In this study, we found that the level of HMGB1 was the highest in Group 1 and the lowest in Group 4. In addition, the level of GAS5 in Groups 1 and 2 was much higher than that in Groups 3 and 4, while the level of miR-1284 in Groups 1 and 2 was much lower than that in Groups 3 and 4. Furthermore, the level of HOTAIR in Groups 1 and 3 was much higher than that in Groups 2 and 4, while the level of miR-22 in Groups 1 and 3 was much lower than that in Groups 2 and 4. Additionally, our results showed that the deletion allele of rs145204276 polymorphism increased the transcription activity of GAS5 promoter, while the G allele of rs4759314 polymorphism increased the transcription activity of HOTAIR promoter. We also found that GAS5 and HOTAIR could directly bind to miR-1284 and miR-22, respectively, while miR-1284 and miR-22 could both target the 3′UTR of HMGB1.
HMGB1 was initially identified as a protein with the chromatin-binding ability. However, HMGB1 has been implicated in the process of cancer cell metastasis, proliferation, and angiogenesis during the development of cancers [Citation11]. In addition, HMGB1 was also shown to play extracellular and intracellular roles by activating critical signalling pathways involved in oncogenesis [Citation11,Citation12]. Moreover, it was demonstrated that the silence of RAGE by siRNA could inhibit HMGB1-induced proliferation of PC cells by reducing the expression of HMGB1 [Citation19]. These studies also showed that the silence of HMGB1 by siRNA could suppress the metastasis of PC cells in an animal model of metastasis.
It has been reported that miR-1284 was down-regulated in osteosarcoma tissues overexpressing HMGB1, indicating the presence of a reverse correlation between the expression of miR-1284 and HMGB1. In addition, miR-1284 can lead to the degradation of HMGB1 by directly binding to its 3′UTR. It was reported that the 3′UTR of HMGB1 contains a binding site of miR-22, and hence the overexpression of miR-22 can suppress the expression of HMGB1 while inhibiting HMGB1-induced autophagy of osteosarcoma cells, and the inhibition of HMGB1 expression by miR-22 in osteosarcoma cells renders the cells more sensitive to the treatments by Cis and Dox by reducing the potential of cancer cell invasion, migration, and proliferation.
Conclusion
In summary, the findings of this study demonstrated the role of HMGB1 in the progression and prognosis of PC. Therefore, the rs145204276 polymorphism located in GAS5 and rs4759314 in HOTAIR may affect the prognosis of PC by modulating the activity of GAS5/miR-1284/HMGB1 and HOTAIR/miR-22/HMGB1 signalling pathways.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386.
- Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol. 2013;64(6):876–892.
- Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem Soc Trans. 2001;29(4):395–401.
- Todorova J, Pasheva E. High mobility group B1 protein interacts with its receptor RAGE in tumor cells but not in normal tissues. Oncol Lett. 2012;3(1):214–218.
- Sims GP, Rowe DC, Rietdijk ST, et al. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28(1):367–388.
- Sparvero LJ, Asafu-Adjei D, Kang R, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7(1):17.
- Kuniyasu H, Chihara Y, Takahashi T. Co-expression of receptor for advanced glycation end products and the ligand amphoterin associates closely with metastasis of colorectal cancer. Oncol Rep. 2003;10:445–448.
- Kuniyasu H, Chihara Y, Kondo H. Differential effects between amphoterin and advanced glycation end products on colon cancer cells. Int J Cancer. 2003;104(6):722–727.
- Caraher EJ, Kwon S, Haider SH, et al. Receptor for advanced glycation end-products and World Trade Center particulate induced lung function loss: A case-cohort study and murine model of acute particulate exposure. PLOS One. 2017;12(9):e0184331.
- Kuniyasu H, Chihara Y, Kondo H, et al. Amphoterin induction in prostatic stromal cells by androgen deprivation is associated with metastatic prostate cancer. Oncol Rep. 2003;10:1863–1868.
- Gnanasekar M, Thirugnanam S, Ramaswamy K. Short hairpin RNA (shRNA) constructs targeting high mobility group box-1 (HMGB1) expression leads to inhibition of prostate cancer cell survival and apoptosis. Int J Oncol. 2009;34:425–431.
- Li T, Gui Y, Yuan T, et al. Overexpression of high mobility group box 1 with poor prognosis in patients after radical prostatectomy. BJU Int. 2012;110(11c):E1125–1130.
- Zhao J, Zhang S, Wu LY, et al. Efficient methods for identifying mutated driver pathways in cancer. Bioinformatics. 2012;28(22):2940–2947.
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799.
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076.
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326.
- Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403.
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208.
- Zhao CB, Bao JM, Lu YJ, et al. Co-expression of RAGE and HMGB1 is associated with cancer progression and poor patient outcome of prostate cancer. Am J Cancer Res. 2014;4(4):369–377.
- Wu T, Zhang W, Yang G, et al. HMGB1 overexpression as a prognostic factor for survival in cancer: a meta-analysis and systematic review. Oncotarget. 2016;7:50417–50427.
- Shen J, Zhang J, Jiang X, et al. LncRNA HOX transcript antisense RNA accelerated kidney injury induced by urine-derived sepsis through the miR-22/high mobility group box 1 pathway. Life Sci. 2018;210:185–191.
- Lv S, Guan M. miRNA-1284, a regulator of HMGB1, inhibits cell proliferation and migration in osteosarcoma. Biosci Rep. 2018;38(4).
- Zhu L, Zhu Q, Wen H, et al. Mutations in GAS5 affect the transformation from benign prostate proliferation to aggressive prostate cancer by affecting the transcription efficiency of GAS5. J Cell Physiol. 2018;234(6):8928–8940.
- Yan H, Zhang DY, Li X, et al. Long non-coding RNA GAS5 polymorphism predicts a poor prognosis of acute myeloid leukemia in Chinese patients via affecting hematopoietic reconstitution. Leuk Lymphoma. 2017;58(8):1948–1957.
- Li Y, Zhao W, Shi R, et al. Rs4759314 polymorphism located in HOTAIR is associated with the risk of congenital heart disease by alternating downstream signaling via reducing its expression. J Cell Biochem. 2018;119(10):8112–8122.
- Geng YJ, Xie SL, Li Q, et al. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39(6):2119–2128.
- Romanuik TL, Wang G, Morozova O, et al. LNCaP Atlas: gene expression associated with in vivo progression to castration-recurrent prostate cancer. BMC Med Genomics. 2010;3(1):43.
- Tao R, Hu S, Wang S, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis. 2015;36(10):1136–1143.
- Zheng Y, Song D, Xiao K, et al. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7(50):83727–83734.
