ABSTRACT
Emergence of multidrug-resistant (MDR) Salmonella enterica serovar Indiana (S. Indiana), a dominant Salmonella serovar in China, has raised global awareness because the MDR S. Indiana also was rapidly emerged in other countries recently. To improve our understanding of underlying MDR mechanism and evolution of this emerging zoonotic pathogen, here we examined the standard ATCC51959 strain together with 19 diverse and representative Chinese S. Indiana strains by performing comprehensive microbiological, molecular, and comparative genomics analyses. The findings from S1-PFGE, plasmid origin analysis and Southern blotting suggested the MDR phenotype in the majority of isolates was associated with large integron-carrying plasmids. Interestingly, further in-depth analyses of two recently isolated, plasmid-free MDR S. Indiana revealed a long chromosomal class I integron (7.8 kb) that is not linked to the Salmonella Genome Island 1 (SGI1), which is rare. This unique chromosomal integron shares extremely high similarity to that identified in a MDR E. coli plasmid pLM6771 with respect to both genomic organization and sequence identity. Taken together, both plasmid and chromosomal integron I exist in the examined MDR S. Indiana strains. This timely study represents a significant step toward the understanding of molecular basis of the emerging MDR S. Indiana.
Introduction
Antimicrobial resistant bacteria pose a significant threat for livestock production, food safety and public health [Citation1]. Recently, the frequent reports of Gram-negative superbugs, particularly those resistant to extended-spectrum cephalosporins, carbapenem, and colistin, have aroused global concern [Citation2]. Salmonella spp. are important foodborne pathogens, to which the elderly, children and immunocompromised people were more susceptible. Antibiotics are still the priority choice for prevention and treatment of salmonellosis because Salmonella spp. usually show a lower resistance rate than other Gram-negative bacteria, such as Escherichia coli and Klebsiella spp [Citation3]. However, the recent emergence of multidrug-resistant (MDR) Salmonella enterica serovar Indiana (S. Indiana) in China has raised global awareness [Citation4].
S. Indiana was first isolated in 1955 from a 9-month-old girl with vomiting, diarrhoea and fever in Indiana, USA. Subsequently, there have been several case reports and outbreaks of S. Indiana-causing vomiting, diarrhoea, fever, gastroenteritis, and abortion in humans and animals in North America and Europe, and sporadic infection reports in the other regions (Africa, South America and Oceania) [Citation4]. In China, since the first isolation of S. Indiana from a foreign visitor in 1984, there have been only six case reports for S. Indiana with a total of 13 isolates being recovered in the subsequent 24 years (1984–2007) [Citation4]. Since 2008, there has been a drastic increase in the number of clinical reports for the infections caused by S. Indiana; notably, the increasing prevalence of S. Indiana in human, animal, food and environment has made S. Indiana become a dominant Salmonella serovar in China, replacing the traditionally dominant serovars, such as S. Typhimurium, S. Derby and S. Agona [Citation4]. More importantly, in parallel to the increased prevalence of S. Indiana, a trend of emergence of highly MDR S. Indiana was also observed [Citation4].
Based on published information, almost all the S. Indiana isolates in China were MDR strains [Citation4]. It is important to mention that nearly 90% of the S. Indiana isolates were resistant to both ciprofloxacin and ceftriaxone/cefotaxime, which are commonly used to treat severe non-Typhi Salmonella infections in adults and children [Citation4]. Moreover, newly identified S. Indiana strains also showed resistance to carbapenem, the last-resort antibiotic for the treatment of MDR infections, suggesting that MDR S. Indiana has become a serious problem [Citation5]. Alarmingly, the MDR S. Indiana was also reported in other countries recently, from Southeast Asia countries to Africa [Citation6–11], suggesting a quick global emergence and expansion of this zoonotic pathogen. Therefore, the MDR S. Indiana is posing an increasing threat to animal health and public health globally.
Extensive studies on other Salmonella serovars have shown that MDR Salmonella strains usually contain unique mobile genetic elements, such as integrons. The integrons were commonly found on conjugative plasmids or in chromosome within the Salmonella Genome island 1 (SGI1) [Citation12]. However, surprisingly, the SGI1 was not identified in S. Indiana based our previous study [Citation13], which indicates the uniqueness of antibiotic resistance (AR) and evolution of S. Indiana when compared to other Salmonella serovars. To date, most S. Indiana studies only focused on the resistance phenotype and the resistance mechanism with respect to the specific drug [Citation4]. Therefore, systematic and genomics study using diverse strains are highly warranted to reveal underlying MDR mechanisms and likely other unique features of S. Indiana [Citation4]. In this study, 19 representative and diverse S. Indiana isolates together with the standard control strain ATCC 51959 were subjected to comprehensive microbiological, molecular, and comparative genomics analyses, which not only revealed wide prevalence of class I integron in the MDR isolates but also discovered a unique long chromosomal integron (7.8-kb) that is not associated with SGI1 as observed in other Salmonella serovars.
Results
Antibiogram of the tested S. Indiana strains
We examined a total of 20 representative S. Indiana strains in this study (). All the S. Indiana strains (), except for ATCC 51959, displayed MDR phenotype with resistance to up to 16 antimicrobials (). In total, 15 resistance patterns were identified and none of the strains was resistant to imipenem and amoxicillin/clavulanic acid (). Based on the number and spectrum of tested antimicrobials, the S. Indiana strains examined in this study were roughly divided into 4 groups (). The Group 1 includes ATCC51959 only, which was susceptible to all tested antimicrobials. The Group 2 includes the four early isolates S0802, S1104, S1105, and S1106 (2008-2011), which exhibited variable resistance to 3–8 commonly used antimicrobials, such as ampicillin, streptomycin, tetracycline, and fluoroquinolones (). Compared to Group 2 strains, the Group 3 showed more consistent resistance to a broader range of antimicrobials with additional new resistance to gentamicin, amikacin and nitrofurantoin (). However, none of the isolates in Group 3 displayed resistance to the third-generation cephems (cefotaxime, ceftriaxone and ceftazidime) and aztreonam. In contrast, the Group 4 isolates, most of which were isolated in 2014 and 2015, not only exhibited resistance to a broad range of antibiotics as group-3 isolates but also showed resistance to the cephems and aztreonam ().
Table 1. The S. Indiana isolates used in this study.
Table 2. Antibiotic resistance phenotype of the tested S. Indiana strains.
Genome mining of MDR determinants
The sequencing coverage and depth were summarized in Table S1 (Supplementary Information). The de novo assembled contigs of the 20 S. Indiana strains were initially aligned against the recently published S. Indiana C629 genome [Citation14]. This initial comparative genome analysis identified limited MDR genes in our S. Indiana strains that displays homology to those present in the C629 plasmid pRCW1 conferring resistance to aminoglycoside, beta-lactam, fluoroquinolone, phenicol, sulphonamide, and trimethoprim [Citation14].
Further comprehensive gene mining using ResFinder [Citation15] for the assembled contigs from each S. Indiana isolates revealed a total of 45 different AR genes in these diverse S. Indiana strains, which may confer resistance to 10 classes of antimicrobials (Table S2). The AR gene profiles of those S. Indiana strains (Table S2) are highly consistent with their resistance phenotypes (). For example, ATCC 51959, which is sensitive to most antibiotics, carries the least extent of AR genes, while S1467 and S1501, which are resistant to the most antibiotics (), carry the broadest spectrum of AR genes (Table S2, Supplementary Information). Interestingly, strain S1318 is susceptible to tetracycline although it carries intact tetA gene and there is no mutation detected in tetA and adjacent regulator tetR, suggesting potential regulatory suppression of tetA in this strain.
Identification and characterization of class I integron in S. Indiana
We observed that a panel of AR genes is clustered in the sequenced S. Indiana genomes. However, the AR gene-bearing plasmid pRCW1 from S. Indiana C629 [Citation14] does not have typical integron due to the lack of conserved signature regions (Data not shown). Thus, simple comparison of the identified AR genes from the 19 S. Indiana strains to the C629 genome cannot help us to link the clustered MDR genes to potential mobile genetic elements in S. Indiana, such as integron.
Interestingly, further database searches revealed that a panel of unique AR gene cassettes in our S. Indiana isolates displayed high level similarity to those in the large plasmid pLM6771 from E. coli 06K2206 (89,090 bp; GenBank accession number: KX009507). Notably, a complete class I integron in pLM6771 was identified using IntegronFinder [Citation16], which is characterized by the presence of one intI1, one attI, three attC sites, and the conserved segment of the integron at 3’ end (3'-CS) comprised of the qacE and sul1 genes (). Other AR genes were also identified within this integron in the pLM6771, which include aminoglycoside N(6)-acetyltransferase gene aac(6’)-Ib, beta-lactamase gene blaOXA-30, chloramphenicol acetyltransferase gene catB3, rifampin ADP-ribosylating transferase gene arr-3 (). In addition, immediately following the 3'-CS, a single copy of insertion sequence common region 1 (ISCR1) gene was identified in pLM6771 (), which encodes a transposase frequently associated with a specific type of class I integron [Citation17]. Finally, additional AR genes adjacent to this integron were also identified in pLM6771, such as those encoding ribosomal RNA methyltransferase ArmA, macrolide efflux protein MsrE, macrolide 2'-phosphotransferase MphE, beta-lactamase IRT-4, beta-lactamase CTX-M-3; the armA, msrE, and mphE genes are depicted in .
Figure 1. Comparison of class I integron in the pLM6771 and the S. Indiana strain S1467. The complete class I integron in S. Indiana S1467 was revealed by filling the gap between the Contig 108 and Contig 33 using primer pair of Ct108 and Ct33 (Table S3; depicted by solid triangles). The grey arrows indicate the class I integron-associated intI1 gene and the typical 3’-conserved segment (3’-CS) containing qacE and sul1 genes. The grey and solid rectangles represent the critical signature sequences of the class I integron (attI and attC sites). The dotted arrow represents the unique transposase gene ISCR1 immediately downstream of the class I integron. The open arrows denote various AR genes (detailed annotations in ).
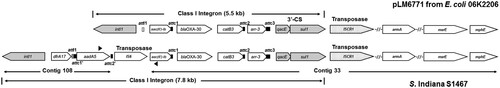
Table 3. Distribution of the major E. coli pLM6771-associated AR genes and mobile elements in different S. Indiana strains.
Strikingly, similar genomic organization of the same AR gene cluster in the E. coli plasmid pLM6771 was directly observed in some assembled S. Indiana genomes, such as S. Indiana strain S1467 and S1501. As shown in , the Contig 108 for S1467 possesses the same signature sequences (intI1, attI, and attC sites) plus additional dfrA17 and aadA5 at the other end; the Contig 33 shares extremely high similarity to the class I integron and its downstream genes carried in pLM6771 with respect to both genomic organization and sequence identity. Using primer pair of Ct108 and Ct33 (Table S3), the gap between Contig 108 and Contig 33 was filled, revealing another transposase gene IS6 (). The IS6 is also located in a short Contig 98 that seems a repeated region. The complete long class I integron identified in S1467 strain (7.8 kb) is depicted in .
In other S. Indiana strains, despite distribution of various AR genes in different contigs, the similar class I integron components and genomic organization of AR genes as shown in were also observed, strongly suggesting that these S. Indiana strains also have similar class I integron. According to our exhaustive genome sequence analysis, distribution of pLM6771-associated AR genes as well as mobile elements in different S. Indiana strains is summarized in . Notably, the core four AR genes within the identified integron, aac(6’)-Ib/blaOXA-30/catB3/arr-3 (), have been discovered in majority of the MDR S. Indiana strains ().
Plasmid analysis: prevalence, origins and location of the class I integron
S1-PFGE showed that plasmids were observed in 16 out of 20 S. Indiana strains, and most of them harboured large plasmids (>200 kb) (). It seems that the strains ATCC51959, S1104, S1467 and S1501 do not carry any plasmids (). However, S1-PFGE approach might miss small DNA fragments (including small plasmids). To further determine if these four strains are indeed plasmid-free, we used two complementary approaches: bioinformatics analysis of plasmid origin and direct plasmid extraction. The assembled contigs from all 20 strains were analysed for plasmid origins using PlasmidFinder [Citation18]. As shown in , the strain S1467 and S1501 did not carry any known plasmid origins, suggesting they are free of plasmids. Furthermore, plasmid extraction using Plasmid Miniprep Kit (Qiagen) did not identify any plasmids in S1467 and S1501 (Figure S1), indicating lack of small plasmids (<20 kb) in these two S. Indiana strains. Therefore, given that both S1467 and S1501 possess most of AR genes () and the complete long class I integron as shown in , the MDR phenotype in S1467 and S1501 should be conferred by the integron located in chromosome. This conclusion is further confirmed by S1-PFGE together with Southern blot analysis. As shown in , Southern blot hybridization using specific probe (targeting qacE) demonstrated that the plasmid-mediated class I integrons were indeed located in the large plasmids of representative S. Indiana isolates (S1319, S1402, S1407, S1459 and S1515), while the integrons in the plasmid-free isolates (S1467 and S1501) were located in chromosome ().
Figure 2. PFGE and S1-PFGE analyses of S. Indiana strains. The left panel is the phylogenetic tree of S. Indiana strains. The middle and right panel is the corresponding PFGE and S1-PFGE result, respectively.
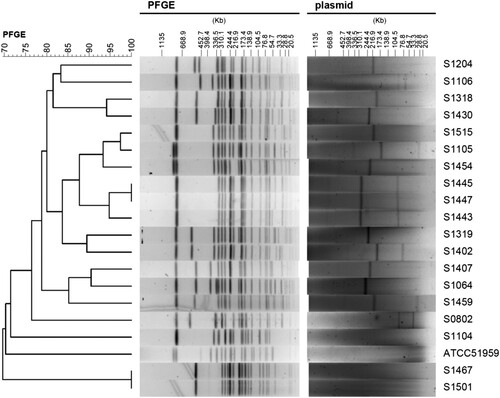
Figure 3. Location of class I integron in S. Indiana strains. S1-PFGE was performed for selected S. Indiana isolates (upper panel). Separated DNA was subsequently transferred into nylon membrane and subjected to Southern blot hybridization against the class I integron by targeting qacE (bottom panel). Lane M, marker H9812; lane 1, ATCC51959; lane 2, S0802; lane 3, S1105; lane 4, S1319; lane 5, S1402; lane 6, S1407; lane 7, S1459; lane 8, S1467; lane 9, S1501; lane 10, S1515.
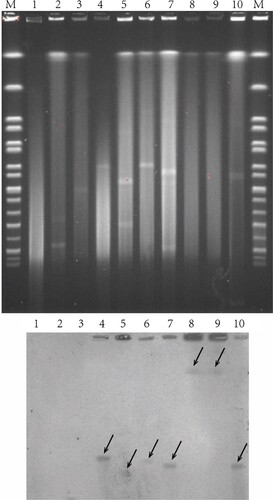
Table 4. Analyses of the plasmids in different S. Indiana strains.
Lack of SGI1 in the S. Indiana strains
It has been widely reported that chromosomal integron was located in SGI1 in Salmonella [Citation12]. However, based on genome sequence analysis in this study, SGI1 was not observed in any of the examined S. Indiana strains. To verify this finding, PCR was performed using a panel of SGI1-specific primers (Table S3, Supplementary Information). Consistently, SGI1 variants were not detected in all the S. Indiana strains including S1467 and S1501 (Figure S2). Therefore, the chromosomal class I integrons in S1467 and S1501 are not associated with SGI1, which is rare in Salmonella. The finding for the lack of SGI1 in S. Indiana in this study, which is supported by whole genome and PCR analyses, also confirmed our previous preliminary observations [Citation13].
Phylogenetic analysis of S. Indiana strains
As shown in Table S4 (Supplementary Information), all S. Indiana strains possess same alleles of the seven housekeeping genes (Salmonella 7-gene dataset from Enterobase), making all the tested S. Indiana strains fall in the same ST17 type based on the MLST analysis. Although MLST typing indicated the S. Indiana strains have a very close phylogenetic relationship, PFGE analysis of the 20 strains () revealed several different patterns. However, some strains, such as S1443 (human strain), S1445 (chicken strain), and S1447 (pigeon strain) which were isolated from different regions, displayed consistent antibiogram, AR gene profile as well as similar PFGE pattern, indicating a potential close or clonal relationship of these isolates. Inconsistency between the phenotype and the genotype was also observed in some strains. For instance, although S1515 and S1105 displayed similar PFGE pattern () and AR gene profile (), unlike S1515, S1105 is a “silent” strain displaying susceptibilities to a panel of antibiotics (), which is likely attributed to an unidentified regulatory mechanism for AR genes in S1105.
Whole genome sequencing typing
Since the genome-scale genetic information is available, whole genome sequencing typing were performed in the PathoBacTyper web server. The whole genome SNP (wgSNP) tree was shown in . The wgSNP tree consists of 3 different clusters. However, the clustering of strains in the wgSNP tree does not seem strictly consistent with the clustering based on resistance phenotype. For example, strain S1105, which is very sensitive to most antibiotics (), is clustered with S1459 and S1454 in wgSNP tree, which displayed MDR phenotype (). However, as observed in PFGE analysis, some strains with similar resistant phenotype were placed in proximity in wgSNP tree, such as S1443/S1445/S1447 and S1467/S1501.
Figure 4. Whole genome SNP tree analysis of S. Indiana strains. Assembled contigs from 20 S. Indiana strains were analysed by PathoBacTyper. The maximum-likelihood phylogenetic tree was constructed with confidence values labelled on the branches. The length of scale bar indicates 1 nucleotide substitution per 100 sites.

Discussion
Currently, there are over 2600 recognized Salmonella serovars. Extensive attentions were placed on those widely prevalent serovars, such as S. Enteritidis and S. Typhimurium, while most Salmonella serovars were considered uncommon serovars. However, some uncommon serovars can arise as prevalent serovars in specific geographic locations [Citation19]. Particularly, recent studies showed that the S. Indiana has evolved from an infrequently reported serovar to one of the most common serovar in China [Citation4]. In parallel to the increased prevalence of S. Indiana, the rapid rise of MDR in S. Indiana also raised serious concerns [Citation4]. Currently, there is a trend of worldwide emergence of the MDR S. Indiana [Citation6–11], highlighting the needs of in-depth characterization of the MDR S. Indiana. In this timely study, to explore molecular mechanisms of MDR S. Indiana, representative Chinese S. Indiana strains were selected for comparative phenotypic and genomic characterizations.
AR gene clusters are of particular interests since they confer resistance to several different antimicrobials or classes of antimicrobials. In this study, we found a unique AR gene cluster (or integron) in diverse Chinese S. Indiana strains; this integron is strikingly similar to the class I integron carried in the plasmid pLM6771 from E. coli 06K2206. According to the submitted information of the pLM6771 genome, the host of this plasmid is a MDR E. coli strain from South Korea. We inferred that the integron observed in this E. coli plasmid is closely related to the integron identified in recent Chinese MDR S. Indiana isolates. However, at this stage, we do not have sufficient epidemiological and genomic evidence demonstrating if this integron was transmitted from S. Indiana to E. coli or from E. coli to S. Indiana. Given that the integron was shared by different bacterial species from different countries, this unique integron may contribute significantly to the emergence and dissemination of MDR globally in Enterobacteriaceae, such as E. coli and Salmonella. This speculation needs to be examined in large-scale studies through international collaboration in the future.
Integrons, enriched with AR genes, can be efficiently disseminated among bacteria via plasmids, particularly with the aid from other mobile elements such as transposon. Previous studies have proposed that MDR phenotype of S. Indiana was likely attributed to the presence of resistant plasmids, including those belonging to IncHI2-type, IncN-type, IncA/C-type, IncP-type and IncFIB-type [Citation13,Citation20,Citation21]. Recently, we observed that a single MDR S. Indiana isolate usually carried one or two plasmids [Citation22] and even 4 plasmids with the presence of resistance genes on 3 plasmids [Citation5]. In this study, S1-PFGE has confirmed this multi-plasmid nature in at least 8 strains ( and ). The Southern blot hybridization demonstrated that the class I integron is located in the large plasmids of all representative plasmid-containing MDR S. Indiana isolates (), suggesting the class I integron is effectively disseminated via plasmid. However, given the presence of the similar integron in diverse plasmids, at this stage, it is still unknown which factors (e.g. transposase, environmental signals) play a critical role in the hopping of the class I integron among different plasmids.
It is of particular interests that two MDR S. Indiana strains (S1467 and S1501) are free of plasmids. Based on plasmid origin analysis and S1-PFGE together with Southern blot hybridization in this study, we obtained compelling evidence indicating the MDR phenotype of S. Indiana strains S1467 and S1501 were conferred by a large chromosomal class I integron. Notably, we also observed that the E. coli pLM6771 belongs to IncL/M group but none of our S. Indiana strains carry either IncL/M or IncA/C plasmid origins (); this finding together with the high similarity of the class I integron in pLM6771 plasmid of E. coli strain to that in the chromosome of S. Indiana S1467 () strongly suggest that the AR gene cluster-containing class I integron can actively hop among plasmids or between plasmid and chromosomes with aid of certain mobile elements, such as the integrase and transposase ISCR1 [Citation17] as shown in . Interestingly, we also demonstrated that the chromosomal class I integron present in these two very recent isolates (S1467 and S1501) was not linked to SGI1, which was rare in Salmonella spp. Since such unique chromosomal integron was not observed in the early MDR S. Indiana, does emergence of this chromosomal integron represent a trend of MDR S. Indiana in the future? This question is highly warranted to be addressed in future large epidemiological investigations, particularly in view of the worldwide spread of SGI1 after the identification of MDR S. Typhimurium DT104 [Citation23].
MLST analysis showed that all the 20 S. Indiana strains belonged to the same MLST type (ST17), which is in agreement with other studies [Citation13,Citation22,Citation24]. According to Salmonella MLST database, S. Indiana isolates in the areas other than China (including France, Denmark, UK and Scotland) also belong to ST17, indicating that S. Indiana exhibited the least sequence types among Salmonella serovars. Despite wide prevalence of S. Indiana in various sources, clearly, the MLST type of S. Indiana is significantly less than those of other prevalent serovars, such as S. Enteritidis and S. Typhimurium. These MLST typing studies all showed that S. Indiana isolates have a very close phylogenetic relationship. Despite many common themes among various Salmonella serovars, presence of a single MLST type for the emerging MDR S. Indiana suggests this specific Salmonella serovar may have acquired unique features to outcompete other serovars during evolution.
To effectively distinguish S. Indiana strains from different sources, the PFGE and whole genome sequencing typing were performed as well for the diverse S. Indiana strains examined in this study. The PFGE result showed that all the 20 S. Indiana strains shared >70% similarity in the PFGE patterns, also the close relationship of the S. Indiana strains. In other reports, S. Indiana isolates also showed high PFGE similarity (50–80%) without predominant PFGE type identified despite diverse phenotypes, which was different from what has been observed in other prevalent serovars such as S. Typhimurium and S. Enteritis [Citation20,Citation22,Citation25,Citation26]. Unlike MLST and PFGE, as expected, whole genome sequencing typing provide unprecedented discriminating power to differentiate highly close lineages. By sequencing the entire genome, most genetic information, such as bacterial classification, AR genes, virulent genes, and SNPs, could be revealed. Besides that, the possible horizontal gene transfer method (via mobile genetic elements) could be analysed. However, the nature of unfinished genomes from high-throughput sequencing still impeded us to get clues for some intriguing findings. For example, although some strains, such as S1515 and S1105, exhibited similar PFGE pattern () and AR gene profile (), they displayed dramatically different AR phenotype (). This interesting finding is likely due to unknown mechanisms of AR gene regulation [Citation27–29]. To test this, obtaining finished genomes of these two isolates are highly warranted in the future.
Although whole genome analysis of the diverse MDR S. Indiana strains in this study has led to novel discovery on molecular basis of the rapidly emerging multidrug resistance in S. Indiana, extensive genome analysis did not help us identify meaningful clues for the evolution of MDR S. Indiana. We did not find unique gene or genomic island that may enable S. Indiana to successfully outcompete the traditionally dominant Salmonella serovars (e.g. S. Typhimurium and S. Derby) although S. Indiana has become a dominant Salmonella serovar in China in the past decade. The MDR S. Indiana also seems to emerge and expand rapidly to other countries based on recent reports [Citation6–11]. Using a univariate and multivariate analysis, recently Collignon et al. [Citation30] proposed that the spread of resistant strains and resistance genes seems to be the dominant contributing factor for the threatening antimicrobial resistance. Therefore, regarding the intriguing S. Indiana, it is likely the MDR S. Indiana acquires significantly enhanced in vivo adaptation during evolution when compared to the other existing MDR Salmonella serovars. To test this, comprehensive assessments of S. Indiana colonization in the host (e.g. chicken) using representative S. Indiana strains and other Salmonella serovar strains are critically needed in the future. Such animal studies in conjunction with in-depth studies using various omics approaches may greatly improve our understanding of S. Indiana evolution and help us develop effective strategies to control the global emergence of MDR S. Indiana.
Materials and methods
S. Indiana strains
A total of 20 representative S. Indiana strains were selected and examined in this study, which include the standard strain ATCC51959 and 19 Chinese isolates (). For these Chinese S. Indiana isolates, 7 isolates (6 from broilers and 1 from poultry worker) was selected from our previous S. Indiana epidemiological study [Citation13], and the remaining 12 isolates was recovered from other sources. To ensure diversity and representativeness, the selection of the 19 Chinese S. Indiana isolates is primarily based on differences in host, isolation year, and geography. Specifically, these diverse S. Indiana strains isolated from multiple sources (), such as human (diarrhoea patients, poultry worker), food (aquatic products, raw chicken) and animal (chicken, duck, goose, pigeon). These S. Indiana isolates are also geographically diverse, which were collected at 16 sites in six provinces in China between 2008 and 2015 ().
Antimicrobial susceptibility testing
All the S. Indiana strains were tested for antimicrobial susceptibility using the disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2013). The S. Indiana strains were tested against 20 antimicrobial agents (OXOID): ampicillin (10 μg), amoxicillin/clavulanic acid (20/10 μg), cefotaxime (30 μg), ceftriaxone (30 μg), ceftazidime (30 μg), aztreonam (30 μg), imipenem (10 μg), streptomycin (10 μg), gentamicin (10 μg), kanamycin (30 μg), amikacin (30 μg), tetracycline (30 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), sulfafurazole (300 μg), trimethoprim (5 μg), sulfamethoxazole/trimethoprim (23.75/1.25 μg), chloramphenicol (30 μg), and nitrofurantoin (300 μg). The E. coli ATCC 25922 was used as a quality control strain.
Pulsed-field gel electrophoresis (PFGE) and Southern blotting
PFGE of the macrorestriction fragment patterns of genomic DNA using XbaI enzyme was performed by following the Centers for Disease Control and Prevention (CDC) standardized PulseNet protocol for Salmonella [Citation31]. S1-PFGE was performed to determine the plasmid profile as described previously [Citation32]. The PFGE profiles of these isolates were clustered by the unweighted-pair group method using average linkages (UPGMA). The isolates whose PFGE patterns displayed a similarity coefficient of >90% were considered as a PFGE cluster having closely related (clonal) genotypes [Citation33]. The location of class I integron was indicated by Southern blot hybridization using a digoxigenin-labelled qacE probe (generated with primer qacE-F/qacE-R by using S. Indiana S1467 genomic DNA as template) according to the manufacturer’s instructions for the DIG-High Prime DNA Labelling and Detection Starter Kit II (Roche Diagnostics, Mannheim, Germany). Ten S. Indiana strains were chosen for Southern blot hybridization, including ATCC51959 (lack of class I integron), S0802 and S1105 (contained incomplete class I integrons), S1319, S1402, S1407, S1459 and S1515 (class I integrons located in plasmids), S1467 and S1501 (class I integrons located in chromosome).
Genome sequencing and comparative genomics analysis
Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen, 69506). The genomic DNA from all the 20 S. Indiana strains were subjected to whole genome sequencing using HiSeq 2500 Sequencing platform at Shanghai Biotechnology Corporation (Shanghai, China), where genomic DNAs were end repaired, ligated to specific adaptors and subject to paired-end sequencing. After filtering raw reads, the clean reads were de novo assembled into contigs using the CLC Genomics Workbench (Genomics Hub, The University of Tennessee, USA). The contigs were aligned against the reference genome of S. Indiana C629 [Citation14] and E. coli 06K2206 plasmid LM6771 (GenBank No: KX009507) by using Mauve (v 2.3.0) [Citation34,Citation35]. S. Indiana C629 was chosen as a template because it was the only S. Indiana strain with full genome sequence available when this project was performed. The assembled contigs from each S. Indiana strains were also uploaded to the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/) for searching acquired AR genes [Citation15] and plasmid origins [Citation18].
Polymerase chain reaction (PCR)
PCR was performed to fill the gap of assembled integron region and to verify the presence of SGI1 variants in S. Indiana. PCR primers were described in Table S3, in which the primers of U7-L12/LJ-R1, 104-RJ/C9-L2 and U7-L12/104-D target the SGI1 left junction, right junction and across junction, respectively [Citation36]. The PCR reaction procedure consisted of an initial denaturation step of heating at 95°C for 5 min, 30 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min. Then there was a final extension step of 72°C for 10 min. The S. Albany strain B0532 (GenBank No: KU288619) and S. Enteritidis standard strain CMCC50041 (GenBank No: CP013097) were used as SGI1 positive control and negative control, respectively.
Multilocus sequence typing (MLST)
Seven conserved house-keeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) were extracted from assembled contigs, and compared with the Salmonella MLST database website (http://mlst.warwick.ac.uk/mlst/dbs/Senterica) to determine the sequence types.
Whole genome sequencing typing
The whole genome sequencing typing was performed using whole-genome-scale single nucleotide polymorphism (wgSNP) method. The assembled contigs and the reference genome C629 [Citation14] were uploaded to the PathoBacTyper server (http://halst.nhri.org.tw/PathoBacTyper/) for performing wgSNP-based genome typing [Citation37].
Nucleotide sequence accession number
The raw sequencing reads of the 20 S. Indiana strains were deposited into GenBank under BioProject PRJNA420683.
Supplementary information accompanies the manuscript on the Emerging Microbes & Infections website http://www.nature.com/emi.
Acknowledgments
JG, CW, and JL designed this study; JG, PZ, and DZ collected the S. Indiana strains, performed antimicrobial susceptibility testing, PFGE analysis and Southern blot hybridization; XZ, CW, and JL performed genome analysis, multilocus sequence typing, whole genome sequencing typing, and analysed the data; JG, XZ, CW, and JL wrote the initial draft of the manuscript; all authors have read and approved the final version of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century – a clinical super-challenge. N Engl J Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651
- Wang Y, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260
- Liu XJ, Lyu Y, Li Y, et al. Trends in antimicrobial resistance against Enterobacteriaceae strains isolated from blood: a 10-year epidemiological study in mainland China (2004–2014). Chin Med J. 2017;130:2050–2055. doi: 10.4103/0366-6999.213407
- Gong J, Kelly P, Wang C. Prevalence and antimicrobial resistance of Salmonella enterica serovar Indiana in China (1984–2016). Zoonoses Public Health. 2017;64:239–251. doi: 10.1111/zph.12328
- Wang J, et al. Complete genetic analysis of a Salmonella enterica serovar Indiana isolate accompanying four plasmids carrying mcr-1, ESBL and other resistance genes in China. Vet Microbiol. 2017;210:142–146. doi: 10.1016/j.vetmic.2017.08.024
- Rayamajhi N, et al. Antibiotic resistance patterns and detection of blaDHA-1 in Salmonella species isolates from chicken farms in South Korea. Appl Environ Microbiol. 2010;76:4760–4764. doi: 10.1128/AEM.02536-09
- Nghiem MN, Nguyen VT, Nguyen TTH, et al. Antimicrobial resistance gene expression associated with multidrug resistant Salmonella spp. isolated from retail meat in Hanoi, Vietnam. Int Microbiol. 2017;20:85–93.
- Nguyen DT, et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int J Food Microbiol. 2016;236:115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017
- Moe AZ, et al. Prevalence and antimicrobial resistance of Salmonella isolates from chicken carcasses in retail markets in Yangon, Myanmar. J Food Prot. 2017;80:947–951. doi: 10.4315/0362-028X.JFP-16-407
- Kiflu B, Alemayehu H, Abdurahaman M, et al. Salmonella serotypes and their antimicrobial susceptibility in apparently healthy dogs in Addis Ababa, Ethiopia. BMC Vet Res. 2017;13:134. doi: 10.1186/s12917-017-1055-y
- Goni AM, Effarizah ME, Rusul G. Prevalence, antimicrobial resistance, resistance genes and class 1 integrons of Salmonella serovars in leafy vegetables, chicken carcasses and related processing environments in Malaysian fresh food markets. Food Control. 2018;91:170–180. doi: 10.1016/j.foodcont.2018.02.039
- Fluit AC. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol Med Microbiol. 2005;43:1–11. doi: 10.1016/j.femsim.2004.10.007
- Gong J, et al. Highly drug-resistant Salmonella enterica serovar Indiana clinical isolates recovered from broilers and poultry workers with diarrhea in China. Antimicrob Agents Chemother. 2016;60:1943–1947. doi: 10.1128/AAC.03009-15
- Wang W, Liu F, Peng Z, et al. Complete genome sequence of Salmonella enterica subsp. enterica serovar Indiana C629, a carbapenem-resistant bacterium isolated from chicken carcass in China. Genome Announc. 2016;4:e00662–16.
- Zankari E, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261
- Cury J, Jové T, Touchon M, et al. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016;44:4539–4550. doi: 10.1093/nar/gkw319
- Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05
- Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14
- Makendi C, et al. A phylogenetic and phenotypic analysis of Salmonella enterica serovar Weltevreden, an emerging agent of diarrheal disease in tropical regions. Plos Negl Trop Dis. 2016;10:e0004446. doi: 10.1371/journal.pntd.0004446
- Bai L, et al. Emergence and diversity of Salmonella enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob Agents Chemother. 2016;60:3365–3371. doi: 10.1128/AAC.02849-15
- Zhang WH, et al. CTX-M-27 Producing Salmonella enterica serotypes Typhimurium and Indiana are prevalent among food-producing animals in China. Front Microbiol. 2016;7:436. doi: 10.3389/fcimb.2017.00436
- Lai J, et al. Unique class 1 integron and multiple resistance genes co-located on IncHI2 plasmid is associated with the emerging multidrug resistance of Salmonella Indiana isolated from chicken in China. Foodborne Pathog Dis. 2013;10:581–588. doi: 10.1089/fpd.2012.1455
- Glynn MK, et al. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901
- Bai L, et al. Prevalence of Salmonella isolates from chicken and pig slaughterhouses and emergence of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana in Henan, China. PLoS One. 2015;10:e0144532. doi: 10.1371/journal.pone.0144532
- Yang B, et al. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shanxi, China. Int J Food Microbiol. 2010;141:63–72. doi: 10.1016/j.ijfoodmicro.2010.04.015
- Lai J, et al. Serotype distribution and antibiotic resistance of Salmonella in food-producing animals in Shandong province of China, 2009 and 2012. Int J Food Microbiol. 2014;180:30–38. doi: 10.1016/j.ijfoodmicro.2014.03.030
- Lowe AM, Beattie DT, Deresiewicz RL. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x
- Cheung AL, Bayer AS, Zhang G, et al. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40:1–9. doi: 10.1016/S0928-8244(03)00309-2
- Zeng X, Brown S, Gillespie B, et al. A single nucleotide in the promoter region modulates the expression of the beta-lactamase OXA-61 in Campylobacter jejuni. J Antimicrob Chemother. 2014;69:1215–1223. doi: 10.1093/jac/dkt515
- Collignon P, Beggs JJ, Walsh TR, et al. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet Health. 2018;2:e398–e405. doi: 10.1016/S2542-5196(18)30186-4
- Ribot EM, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59
- Li R, Xie M, Lv J, et al. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother. 2017;72:696–699.
- Sahin O, et al. Molecular evidence for zoonotic transmission of an emergent, highly pathogenic Campylobacter jejuni clone in the United States. J Clin Microbiol. 2012;50:680–687. doi: 10.1128/JCM.06167-11
- Darling AC, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704
- Rissman AI, et al. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356
- Boyd D, Cloeckaert A, Chaslus-Dancla E, et al. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob Agents Chemother. 2002;46:1714–1722. doi: 10.1128/AAC.46.6.1714-1722.2002
- Tsai MH, Liu YY, Soo VW. Pathobactyper: a web server for pathogenic bacteria identification and molecular genotyping. Front Microbiol. 2017;8:1474. doi: 10.3389/fmicb.2017.01474
