ABSTRACT
Recent extensive (re)emergences of arthropod-borne viruses (arboviruses) such as chikungunya (CHIKV), zika (ZIKV) and dengue (DENV) viruses highlight the role of the epidemic vectors, Aedes aegypti and Aedes albopictus, in their spreading. Differences of vector competence to arboviruses highlight different virus/vector interactions. While both are highly competent to transmit CHIKV (Alphavirus,Togaviridae), only Ae. albopictus is considered as a secondary vector for DENV (Flavivirus, Flaviviridae). Among other factors such as environmental temperature, mosquito antiviral immunity and microbiota, the presence of non-retroviral integrated RNA virus sequences (NIRVS) in both mosquito genomes may modulate the vector competence. Here we review the current knowledge on these elements, highlighting the mechanisms by which they are produced and endogenized into Aedes genomes. Additionally, we describe their involvement in antiviral immunity as a stimulator of the RNA interference pathways and in some rare cases, as producer of viral-interfering proteins. Finally, we mention NIRVS as a tool for understanding virus/vector co-evolution. The recent discovery of endogenized elements shows that virus/vector interactions are more dynamic than previously thought, and genetic markers such as NIRVS could be one of the potential targets to reduce arbovirus transmission.
The main vectors of many medically important arboviruses, such as chikungunya (CHIKV), zika (ZIKV) and dengue (DENV) viruses, are the two mosquito species Aedes aegypti and Aedes albopictus. While their extensive distribution covering most tropical, subtropical and even, temperate countries, makes them a real threat for human health, Ae. aegypti and Ae. albopictus have different historical backgrounds and do not exhibit the same efficiency to transmit arboviruses. The objectives of this review are to point out critical features of both mosquito species that could explain their differences in vector competence. Vector competence is modulated by environmental, genetic, and epigenetic factors, the latter including mechanisms induced by mosquito microbiota [Citation1]. Recently, non-retroviral integrated RNA virus sequences (NIRVS) have been proposed to be among the genetic factors influencing vector competence. The potential role of NIRVS in mosquitoes as vectors is discussed.
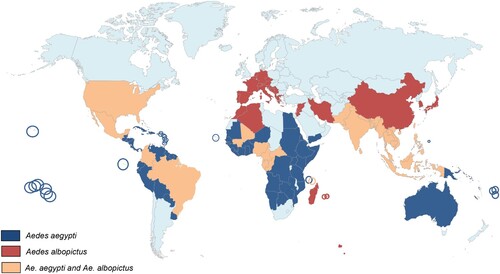
Aedes albopictus and Aedes aegypti have different histories ()
Aedes albopictus (Skuse, 1894) is a mosquito species closely related to Ae. aegypti, both belonging to the Culicidae family and vectors of several different arboviruses highly pathogenic for humans such as chikungunya virus (CHIKV) [Citation2,Citation3], yellow fever virus (YFV) [Citation4] and dengue viruses (DENV) [Citation5,Citation6]. Contrary to many other mosquito vectors such as the malaria vector Anopheles gambiae, Ae. albopictus and Ae. aegypti eggs are capable of entering in diapause and quiescence respectively, ensuring survival during and after environmental stress [Citation7–10]. In addition to survive under extreme conditions, this characteristic allows the two vectors to colonize new regions around the world [Citation11].
However, in terms of evolution, the two species have a different history. Aedes aegypti (Linné, 1862) originates from a sub-Saharan African sylvan ancestor that migrated to West Africa late in the 8th century. It was introduced in the New World probably via the African slave trade between 15th and 17th centuries [Citation12,Citation13]. Around 1800, the species was introduced in the Mediterranean region where it was established in European harbours until about 1950 [Citation14]. Aedes aegypti was introduced into Asia from Europe with the opening of the Suez Canal in 1869; it is abundantly found in Asia since late nineteenth century [Citation15]. The species was later introduced in Australia (1887) and the South Pacific (1904) [Citation14]. On the other hand, Ae. albopictus is native to tropical forests of South-East Asia. Until the late 70s, this species was restricted to Asia, India and a few islands in the Pacific region such as La Reunion [Citation16], the Seychelles [Citation17] Mariana and Papua New Guinea islands [Citation18]. However, in less than three decades, it has conquered all continents except Antarctica [Citation19,Citation20]. Contrary to Ae. aegypti which took hundreds of years to cover the tropical world, Ae. albopictus took only few decades to wide spread. This impressive fast colonization, promoted by increased human mobility and trade of goods including used tires and lucky bamboo as potential mosquito breeding sites, stresses its high ability to survive under both tropical and temperate regions. Moreover, Ae. albopictus is also a serious threat for human populations as it is a competent vector for at least 26 different arboviruses [Citation21] and filarial nematodes of veterinary and zoonotic significance [Citation22,Citation23].
Both species are involved as vectors in major human diseases ()
After suspected outbreaks in America and Asia in the 18th and 19th centuries [Citation24], CHIKV has been first identified in Tanzania in 1952 where it circulated between non-human primates and mosquito vectors. The virus escaped from a sylvatic cycle to cause urban outbreaks in South East Asia and Africa from the 1960s with Ae. aegypti as the main vector (reviewed by [Citation25]). Lastly, CHIKV re-emerged in Thailand in 1991 [Citation26], in the Democratic Republic of Congo in 1999–2000 [Citation27], then in coastal Kenya in 2004 [Citation28], and in the Union of Comoros, in 2005 [Citation29,Citation30], mainly associated to Ae. aegypti. More recently, the same species was involved in CHIKV outbreaks in 45 countries and territories in America, causing almost 3 million cases from 2013 to 2016 (https://www.paho.org/hq/dmdocuments/2014/2014-jun-20-cha-CHIKV-authoch-imported-cases-ew-25.pdf; https://www.paho.org/hq/dmdocuments/2015/2015-sep-18-cha-CHIKV-cases-ew-37.pdf). On the other hand, Ae. albopictus was also proved to be susceptible to CHIKV infection [Citation31] and could involve as a CHIKV vector. In 2005, it became the primary vector on La Réunion Island where Ae. aegypti was present as remote populations [Citation32–34]. On this island, CHIKV acquired a mutation in the glycoprotein E1 (E1-A226V) [Citation35] that increases its infectivity in Ae. albopictus but not in Ae. aegypti [Citation36,Citation37]. From there, the virus spread to India and Southeast Asia between 2007 and 2014 causing 1.4 million cases [Citation38].
Table 1. List of arboviruses transmitted by Aedes aegypti and Aedes albopictus.
Aedes aegypti and Aedes albopictus are also vectors of DENV, with the former being the major vector. Aedes aegypti has been responsible for severe outbreaks in America, Southeast Asia and Western Pacific regions in the late twentieth century [Citation39,Citation40]. On the other hand, even though it was also responsible for severe DENV outbreaks such as during the World War II in Japan [Citation41] or recently in China [Citation42], Ae. albopictus is considered as a less efficient vector of DENV. Indeed, no major epidemics were reported in regions like Taipei, Guam or Hawaii islands where Ae. albopictus is predominant, even when nearby places suffered from DENV outbreaks involving Ae. aegypti (reviewed by [Citation43]). Additionally, even in presence of DENV outbreaks due to Ae. albopictus, like in the Seychelles Islands (1977), La Réunion Island (1977), southern China (1978), Macao (2001), Hawaii (2001) and lastly, in Europe [Citation44,Citation45], only mild symptoms are described [Citation46].
Aedes albopictus and Aedes aegypti have different vector competences
To be a vector, the arthropod species must be competent. The vector competence is the ability of an arthropod to acquire, support replication and dissemination of an infectious agent and successfully transmit it to another susceptible host [Citation47,Citation48]. Vector competence is a component of vectorial capacity and is determined by both genetic (depending on mosquito species/population, virus genotype/strain and their interactions) [Citation49] and non-genetic factors (e.g. environmental components) [Citation50]. Aedes albopictus and Ae. aegypti are highly susceptible to different CHIKV strains (Table S1) (reviewed by [Citation51]). Although Ae. albopictus is considered as a secondary vector for DENV [Citation21], its susceptibility to DENV infection compared to Ae. aegypti remains controversial [Citation52–56]. Aedes albopictus mosquitoes generally show a higher midgut susceptibility to DENV infection but a lower rate of virus dissemination compared to Ae. aegypti (Table S2) [Citation43].
Epigenetic factors which include mechanisms associated with the vector microbiota contribute to vector competence [Citation1]. Insect microbiota comes from the environment: the breeding sites where immature stages live [Citation57] and the flower nectar where adults get the sugar nutrient as carbon source [Citation58]. Insect microbiota influences various physiological processes that favour insect ecological adaptation such as growth, reproduction, survival and tolerance to external stresses [Citation59–63]. Moreover, insect microbiota is capable of stimulating immune responses, described as immune priming, conferring antiviral protection [Citation64,Citation65]. As an example, infection with the bacterium Wolbachia induces an oxidative stress in Ae. aegypti causing an increased level of reactive oxygen species (ROS). The elevation of ROS activates immune pathways, inhibit DENV and then affect the vector competence [Citation66].
Insect immunity: Toll, Imd, JAK-STAT pathways
The antiviral role of the microbiota has been ascribed to the activation of immune pathways [Citation66,Citation67]. So anti-viral immunity in mosquito vectors is critical to prevent virus replication and transmission. Mosquitoes lack adaptive immune responses, but they present innate immunity based on several strategies such as encapsulation and phagocytosis, melanization and production of physical barriers. Moreover, several molecular pathways have been described with antiviral immunity activities, including the RNA interference (RNAi) system, discussed further in the review, the Toll, Immune deficiency (Imd), Janus Kinase-Signal Transduction and Activators of Transcription (JAK-STAT) pathways [Citation67–72]. The activation of these pathways leads to the expression of effector genes that have antiviral activities.
The Toll pathway of mosquitoes is very similar to the mammalian Toll-Like Receptor pathway (TLR). This pathway is activated by the interaction between either viral pathogen-associated molecular patterns (PAMPs) or the putative Toll ligand Spätzle, and host pattern recognition receptors (PRRs), that are present in several parts of the body (hemocele and midgut). This interaction leads to the recruitment of Myd88 that triggers the phosphorylation and degradation of the negative regulator Cactus and the nuclear translocation of the NF-kB-like transcription factor Rel1 that induces the transcription of antimicrobial peptides, such as cecropins and defensins [Citation67].
The first (PRR activation) and the final step (synthesis of antimicrobial peptides) of the Imd pathway are processed in the same way as the Toll pathway, but different intermediate components are required in the cascade of the signalling pathway. The NF-kB-like transcription factor Rel2 is activated by the caspase-mediated cleavage and is then translocated to the nucleus where it triggers the transcription of Imd-related genes [Citation73].
The JAK-STAT pathway is activated through the interaction between the Unpaired ligand (Upd) and the receptor Dome. It first promotes the binding of Janus kinases to Dome and then the recruitment of STAT proteins. Once activated, STAT proteins are translocated into the nucleus and trigger the transcription of antimicrobial related genes, and specific antiviral genes such as vir-1 (virus-induced RNA 1) [Citation74–76].
Studies in Ae. aegypti revealed that the Toll and JAK-STAT pathways were both upregulated 10 days after DENV infection suggesting an anti-DENV activity [Citation67,Citation77]. Moreover, the JAK-STAT pathway has an antiviral activity against another flavivirus, WNV in Culex mosquito cells [Citation71]. However, although suggested in Drosophila [Citation69,Citation78], antiviral properties of these signalling pathways are less obvious for the alphaviruses of the Togaviridae family. In both in vitro and in vivo experiments with Ae. aegypti , none of the 3 above-mentioned pathways showed anti-CHIKV properties [Citation79]. Additionally, in Ae. albopictus-derived U4.4 cells, infections with the Alphavirus Semliki Forest virus did not trigger the JAK/STAT, Toll and Imd pathways [Citation80]. Primed by the mosquito microbiota, the Imd pathway showed antiviral effects in Ae. aegypti following a blood meal containing the Alphavirus Sindbis virus (SINV) [Citation81]. Ultimately, a microarray analysis on Ae. aegypti infected by SINV revealed a temporary up-regulation of Toll pathway which was later inhibited by the virus [Citation82]. Collectively, these results suggest that antiviral immunity in mosquitoes is in part controlled by the Toll, Imd and JAK-STAT pathways which are efficient against flaviviruses, such as DENV and WNV, but their action on alphaviruses such as CHIKV and SINV is less obvious suggesting a virus-specific antiviral regulation.
Genome characteristics and evolution
Quantitative trait loci (QTL)
Because vector competence is under the control of multiple genes, quantitative genetics have been used to measure the contribution of mosquito genetic factors to viral infection and dissemination in mosquitoes. Quantitative Trait Loci (QTL) are defined as several genes grouped in the genome that affect the expression of quantitative traits and lead to important phenotypic variations. The species Ae. aegypti is described under two forms: Ae. aegypti formosus for the ancestral African type breeding in tree holes and Ae. aegypti aegypti for the domestic type colonizing man-made containers [Citation83]. Using intercrosses of Ae. aegypti aegypti and Ae. aegypti formosus strains, respectively highly and weakly susceptible to DENV infection, two QTLs were identified: one affecting the midgut infection barrier on chromosomes 2 and 3, and one on chromosome 3 associated with a midgut escape barrier [Citation84]. Moreover, an additional QTL found on the chromosome 2 along with a sex-linked QTL were associated with the ability to infect the midgut [Citation85]. Moreover, QTLs were identified on the 3 chromosomes of Ae. aegypti associated with DENV-2 dissemination from midguts [Citation86]. It appears that several different parts of the Ae. aegypti genome identified as QTL are independently capable of modulating the vector competence to DENV-2. However, no studies to date have been conducted to identify potential QTLs affecting the vector competence to DENV in Ae. albopictus genome.
Transposable elements (TE)
Last technical improvements in genome sequencing allowed bringing to light the complexity of mosquito genomes. Aedes mosquitoes have the biggest genome size among currently-sequenced mosquito genomes. For instance Ae. aegypti genome is 1,380 MB; [Citation87], Ae. albopictus is 1,900 MB [Citation88,Citation89] while the Anopheles gambiae genome is 278 MB; [Citation90] and Culex quinquefasciatus is 579 MB; [Citation91].
Differences observed in the genome size of Ae. albopictus could be explained by the presence of Transposable Elements (TEs) [Citation92,Citation93]. First discovered in 1956 [Citation94], TEs are considered as intragenomic parasites [Citation95,Citation96]. Ubiquitously found in both prokaryotic and eukaryotic genomes, TE are described as sequences integrated in the host genome capable of both independent replication and movement from one chromosomal location to another through a phenomenon called transposition. Transposition can occur in both somatic and germ line cells. However, some elements transpose in specific cell types, like the P elements in Drosophila melanogaster [Citation97] or without any cells preference, such as the bacteriophage Mu [Citation98,Citation99]. Transposons are classified into two groups, depending on their DNA structure and transposition mechanism. The class I, also called retrotransposons, relies on RNA intermediates to transpose and is divided in two subgroups: LTR (Long Terminal Repeats) retrotransposons and non-LTR retrotransposons (reviewed by [Citation100]). The class II TEs, also called DNA elements, contains terminal inverted repeats (TIRs). Three different groups of DNA elements have been described in eukaryotes: classic transposons [Citation101], helitrons [Citation102] and mavericks, also called politons [Citation103]. Unlike retrotransposons, DNA elements do not rely on RNA intermediates for transposition [Citation104].
Transposons are major drivers of host genome function and evolution. They can act as a source of mutational variations through their transposition producing multiple copies of the same element in the host genome. These copies can facilitate regulation of gene expression, recombination and unequal crossing-overs between chromosomes and therefore, lead to chromosomal rearrangements by creating deletions, insertions, duplications, inversions and translocations. When a TE insertion occurs in an exon, the ORF can change and codes for a non-functional peptide or cause missense or nonsense mutations. A TE insertion can also create alternative splicing leading to the production of several protein isoforms or introduce a polyadenylation signal [Citation105,Citation106]. TE activity in a host genome contributes to introduce diversity. In Ae. albopictus genome, the differences of genome size are explained by the amount of TEs which represents 68% (1,967 Mb) of the total genome [Citation89]. Additionally, variations of repetitive sequences were detected at the intra- and interspecific levels [Citation88,Citation92,Citation107,Citation108]. When comparing the TE composition between Ae. albopictus and Ae. aegypti, differences in the quantity and type of repeats are seen; TE amount reaches 1,343 and 988 Mb in the Ae. albopictus and Ae. aegypti genomes, respectively [Citation89]. More than 20% of repetitive sequences present in Ae. albopictus are absent in Ae. aegypti. The two species have diverged 71 million years ago and most TE insertions occurred during the last 10 million years in the Ae. albopictus genome [Citation89]. DNA transposons represent only 8% of TEs present in the Ae. albopictus genome, and 15% in the Ae. aegypti genome [Citation89]. Non-LTR retrotransposons LINE represent one third of TEs in both genomes, followed by a high proportion of LTR retrotransposons, suggesting that retrotransposons and DNA transposons are suspected to cause genome size variations between Ae. aegypti and Ae. albopictus. Moreover, the activity of TEs can be controlled by the siRNA and piRNA immune pathways. piRNAs and siRNAs produced respectively by TEs from class I and class II transposons, can be up-regulated after an infectious blood feeding leading to modify the outcome of infection, and then the vector competence [Citation109].
Endogenous viral elements (EVEs)
Due to strong and long-lasting interactions between the virus and the vector, the virus could integrate whole or parts of its genome into the genome of host cells, leading to the formation of Endogenous Viral Elements (EVEs) [Citation110]. These elements are defined as viral sequences that integrate into the host germline as double-stranded DNA and are therefore maintained in the population through vertical transmission to the progeny. Considering that the genome of germline cells are strongly protected against any kind of intrusions, such as TE activity, notably by piRNAs [Citation111], the odds of EVE introduction must be low. However, around 7%–8% of the human genome is made up by sequences of viral origins [Citation112].
EVEs originated from retroviruses are called Endogenous Retroviruses (ERVs). It is well known that ERVs formation occurs frequently in host cells since the integration into the genome host cell is mandatory to complete their viral life cycle. ERVs are easily detectable because of Long Terminal Repeats (LTR) present at each end of the segment. Other EVEs originated from other viral families have been recently discovered in many host genomes: single-stranded DNA viruses such as Circoviridae and Parvoviridae in diverse vertebrate genomes (dog, mouse and panda; [Citation113]) and double-stranded DNA viruses such as hepadnaviruses in zebra finch genome [Citation114].
Non-retroviral integrated RNA virus sequences (NIRVS) ()
Main characteristics
Since non-retroviral RNA viruses do not encode for reverse transcriptase or integrase, endogenous enzymes or viruses infecting the cell at the same time must be involved in the endogenization of such viruses into host genome DNA. Three steps should be involved to achieve the integration of non-retroviral RNA viruses into the host genome: (i) first, the non-retroviral RNA needs to be reverse-transcribed into viral-derived double-stranded DNA (vDNA), (ii) be imported in the nucleus, and (iii) finally be integrated into the host genome.
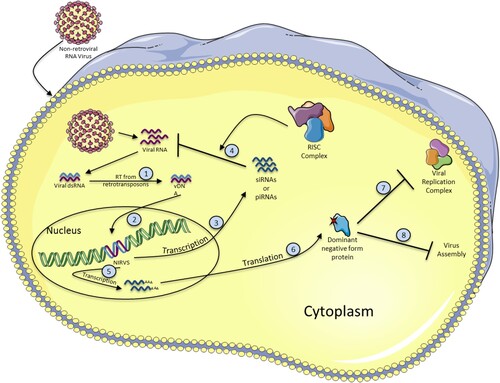
Figure 2. Formation and antiviral functions of NIRVS. When a non-retroviral virus infects a cell, the viral RNA is released and double stranded RNA (dsRNA) intermediates are produced. Viral dsRNA is then used as a template to produce viral DNA (vDNA) by the reverse transcriptase activity of retrotransposon elements (1). vDNA integrates into the host cell genome, probably by transposition activity of retrotransposons, becoming a NIRVS (2). NIRVS is then transcribed either into siRNAs or piRNAs (3) to inhibit the viral RNA after association with the RISC complex (4) or into mRNA (5), and translated into a dominant negative form protein (6), that can alter the viral replication by several ways. For example, by inhibiting the viral replication complex (7) or viral assembly (8).
The first mosquito NIRVS were identified in 2004 in Aedes spp. cell lines and mosquitoes [Citation115]. Most of them were truncated or incorporated several stop codons, but one contained an intact ORF homologous to the NS1-NS4A region of insect-specific viruses (ISVs) in Ae. albopictus genome, i.e. Cell Fusing Agent Virus (CFAV) and Kamiti River Virus (KRV). This last fragment represents around one half of the flaviviral genome. These NIRVS (also called Cell Silent Agent (CSA) sequences) comprised two third of the flaviviral genome and contained enzymatic domains such as helicase and serine protease. The corresponding mRNA was detected in C6/36 Ae. albopictus cells suggesting the expression of the NIRVS and its potential functional role in the cell at the RNA level since no protein was detected [Citation116]. Moreover, this NIRVS is present in 97%–98% of Ae. albopictus mosquitoes. Many NIRVS were found homologous to insect-specific flaviviruses (ISFs), such as CFAV, KRV and Aedes Flavivirus (AeFV) closely related to arboviruses [Citation117,Citation118]. The high prevalence of NIRVS in Aedes spp. genome as well as the high frequency of transposons might somehow be correlated to the mosquito genome size [Citation87,Citation89].
Most NIRVS described up to date were found in Aedes spp. genomes. Among 424 RNA viruses detected in 22 mosquito genomes, 81% (194/239) were identified as NIRVs in Aedes genomes and among them, 63% of NIRVS were located into the Ae. aegypti genome, and the remaining 37% were identified in Ae. albopictus [Citation119]. Additionally, 72% of the NIRVS were homologous to the Rhabdovirus family whereas 27% were close to the Flavivirus genus and 1% left belonging to Bunyavirus and Reovirus genera. These data are consistent with another study which compared the “EVEome” of both Ae. aegypti and Ae. albopictus [Citation120]. Factors leading to endogenisation of viral genomes into host cells remain unclear. The mRNA abundance could be critical since ssRNA+ genomes are directly translated into proteins and ssRNA- genomes have first to be transcribed. Moreover, the transcripts of flaviviral genome are usually longer than those from ssRNA- viruses, and this could decrease their chance to be integrated into the host genome [Citation121]. Most of the flaviviral NIRVS detected in silico are originated from non-structural protein coding sequences rather than structural ones. In Ae. aegypti and Ae. albopictus genomes, 30 and 25 flaviviral NIRVS were mapped to non-structural protein coding sequences, and respectively, only 2 and 3 NIRVS represented similarities with structural proteins coding sequences [Citation119]. Half of the rhabdoviral NIRVS mapped to the N gene, which encode the nucleoprotein [Citation110,Citation122]. From 3’ to 5’, each gene (N, P, M, G and L) of the rhabdoviral genome is transcribed in a progressive graduated manner due to the recognition of stop codons/polyadenylation signals by the polymerase [Citation123], meaning that the transcripts at the 3’ end (i.e. N gene) are in higher quantities than for those near the 5’ end (i.e. L gene).
Production of viral DNA (vDNA) from non-retroviral viruses
As previously mentioned, to become integrated into the host genome, the non-retroviral RNA virus is first reverse transcribed to produce viral DNA (vDNA), imported into the cell nucleus and finally integrate into the chromosome [Citation124–127]. Interestingly, only some parts of the viral genomes can be found in a DNA form. The reverse transcriptase probably switches from the original RNA template to a close viral RNA genome causing multiple independent reverse-transcription events [Citation128,Citation129]. These vDNA could also be the result of replication-slippage events caused by the reverse transcriptase. Whether the vDNA form belongs to the host genome or is present as extra-chromosomal DNA element such as episomes is still unknown. RNAi-deficient cells (C6/36) possess more vDNA forms than RNAi-proficient cells (Aag2 cells) suggesting that RNAi system could inhibit vDNA production. More importantly, after mosquito infection with CHIKV, vDNA has been found in legs and wings of infected Aedes mosquitoes suggesting that either vDNA is capable of dissemination from one tissue to another (possibly through cellular and tissue damages) in the mosquito or that all infected cells produce vDNA [Citation125]. Moreover, FHV and Sindbis vDNA were found in infected flies after infection [Citation130].
NIRVS reverse transcription and integration mediated by retrotransposons
vDNA from DNA viruses can integrate into host chromosomes by Non-Homologous (double-stranded) End Joining (NHEJ) [Citation114,Citation131], Non Homologous DNA recombination used by adeno-associated DNA virus [Citation132–134] or Telomeric homologous recombination [Citation135]. However, little is known about the mechanism used by the NIRVS to integrate into host chromosomes. Nevertheless, reverse transcription activity from endogenous retrotransposons has been associated with vDNA formation [Citation130]. By adding a reverse transcriptase inhibitor, azidothymidine (AZT) in S2 and Kc167 Drosophila cell cultures, vDNA formation was inhibited after infection with several RNA viruses, namely Flock House Virus, Sindbis Virus and Drosophila C Virus (DCV) [Citation130]. vDNA of CHIKV and DENV were detected after infections in Ae. albopictus and Ae. aegypti mosquitoes and cell cultures [Citation125]. vDNA plays an important role in viral tolerance rather than viral resistance [Citation125]. The early production of vDNA (6 hours and 2 days post-infection in cultured cells and mosquitoes respectively) is critical to establish efficient immune responses [Citation125]. These regions were also enriched with LTR retrotransposons as it has been shown in Drosophila, especially retrotransposons of the Ty3_gypsy and Pao Bell families [Citation119,Citation120]. This suggests an important role of LTR retrotransposons in the reverse transcription of vDNA.
Biological function of NIRVS
The integration of NIRVS into host genomes has now been recognized to occur more frequently than previously thought. It has been suggested that NIRVS could be involved in antiviral immunity [Citation128,Citation136]. A non-retroviral RNAs segment encoding the capsid protein of the Israeli Acute Paralysis Virus (IAPV), a ssRNA+ dicistrovirus, was found in the genome of one third of the honeybee population (Apis mellifera); it was correlated with a virus-resistant phenotype [Citation137]. Moreover, the presence of vDNA allowed the survival of FHV-infected flies [Citation125,Citation130]. More precisely, vDNA production detected at early stages of infection, promoted viral persistence, as it has been seen in in vivo and in vitro experiments with mosquitoes challenged with CHIKV and S2 FHV-infected Drosophila cells [Citation125,Citation130].
The antiviral function of NIRVS has been linked to the innate immune system of RNAi which has been shown to be the main antiviral system in insects [Citation125,Citation138,Citation139]. This system relies on small RNAs (sRNA) that when associated with a complex of proteins recognized by sequence-complementarity, led to the cleavage and degradation of incoming foreign nucleic acids [Citation140]. Three different pathways have been described: the small interfering RNA (siRNA), the micro RNA (miRNA) and the PIWI-interacting RNA (piRNA). All three use the same mechanism to perform their antiviral action, but are distinguished by the sRNA biogenesis and the protein complex involved. Whereas the role of siRNA pathway in viral immunity in mosquitoes is largely accepted, little was known about the function of the piRNA pathway except its role in preserving genome stability in the germline by regulating the activity of transposable elements in D. melanogaster [Citation141–145] and Aedes mosquitoes [Citation146,Citation147]. However, the piRNA pathway has been linked to antiviral immunity both in vitro and in vivo [Citation148–153]. Indeed, deep-sequencing analysis of DENV-2 infected Ae. aegypti Aag2 cells revealed the production of specific viral piRNAs (vpiRNAs) along with viral siRNAs (vsiRNAs) [Citation154]. Moreover, vpiRNAs have been detected in DENV-infected Ae. aegypti individuals as early as 2 days post-infection [Citation151]. Nevertheless, the piRNA pathway has no antiviral property in the insect model D. melanogaster suggesting a different function depending on the host [Citation155].
Interestingly, EVEs including NIRVS present in Aedes mosquitoes are frequently located in TE-derived piRNA clusters [Citation119,Citation120]. In Ae. aegypti and Ae. albopictus, half of NIRVS mapped to piRNA clusters in Ae. aegypti genome and only 12.5% of NIRVS mapped to piRNA clusters in Ae. albopictus genome [Citation119], suggesting that the presence of NIRVS in these clusters was not a general feature. Moreover, bioinformatic predictions on Aag2 cell line showed that piRNA clusters containing EVEs produced more piRNA than those without EVEs, meaning that viruses may not integrate randomly in the host genome but target specific active piRNA clusters for endogenization [Citation120]. Additionally, NIRVS produced both primary and secondary piRNAs; immunoprecipitation of Piwi proteins also detected NIRVS-derived sRNAs, and knockdown of Piwi proteins resulted in a decrease of NIRVS-derived sRNA expression [Citation119]. However, NIRVS-derived siRNAs were not found indicating that NIRVS are involved in only one specific RNAi pathway. NIRVS originated from insect-specific viruses were proved to produce antisense orientation primary piRNA-like molecules and be located in active regions of both siRNA and piRNA production in Ae. aegypti and Ae. albopictus mosquitoes [Citation116]. In CHIKV-infected Ae. aegypti and Ae. albopictus, NIRVS produced viral small-interfering RNAs (vsiRNAs) and probably vpiRNAs after infection [Citation125]. In FHV-infected Drosophila cells treated and non-treated with AZT (inhibitor of reverse transcriptase), vDNA are transcribed and processed by the RNAi machinery into vsiRNAs [Citation130]. The knocking-down of RNAi machinery in Drosophila infected cells resulted in an acute infection leading to cell death [Citation130].
In summary, NIRVS located in specific regions of the genome such as TE-derived areas called piRNA clusters in mosquitoes, are important for RNAi-based immunity [Citation156]. Their transcripts are capable of producing vsiRNAs in Drosophila [Citation130] and both vsiRNAs and vpiRNAs in Aedes mosquitoes [Citation119,Citation120,Citation125]. The production of sRNAs is induced following arboviral infections (Togaviridae and Flaviviridae) and NIRVS are required for mosquito tolerance to control viral infection [Citation125]. Since vDNA has been found in many mosquito tissues following viral infection, vDNA could serve as a danger signal to warn the uninfected cells and implement a solid immune response through sRNA production [Citation125], even though the virus could also counteract by producing VSR (Viral Suppressor of RNAi), as it has been seen with insect-specific viruses [Citation157,Citation158].
NIRVS functional role at the protein level
Even though some NIRVS have accumulated several mutations including stop codons, some of them have conserved their open reading frames (ORFs) suggesting that they could be translated into proteins and have a function at the protein level. This scenario was first described for many Endogenous Retroviral elements (ERVs) found in different host genomes [Citation159]. Produced proteins can confer viral interference and direct antiviral properties, leading to a resistance phenotype [Citation160–162]. Up to now, no biological functions were found at the protein level for NIRVS in mosquitoes. However, many of them were proved to produce transcripts, mostly in Aedes and Anopheles mosquitoes [Citation115,Citation116,Citation119,Citation122,Citation163] meaning that related proteins should be discovered shortly. Collectively, these results suggest that NIRVS have biological functions rather than being endogenized randomly into host genomes. Despite their low or even undetectable levels of RNA [Citation119], NIRVS are suggested to be involved in the main antiviral defense mechanism in mosquitoes as being a source of sRNA production [Citation116,Citation119,Citation120,Citation125,Citation130]. In some rare occasions, NIRVs produce a protein which blocks viral infection and replication by affecting viral polymerase activity [Citation161].
NIRVS as ancient scars attesting virus/host coevolution
Understanding ancient viral cross-species transmission events and how viruses have evolved and interacted with their hosts in the past is important for anticipating future emerging diseases. However, reconstituting the history of viruses remains a challenge considering their rapid evolution. Indeed, viruses are considered as the fastest-evolving biological entity with an evolution rate of 10−3 substitutions/site/year (s/s/y) [Citation164–167]. Once endogenized in the host, NIRVS are submitted to a slower evolution rate, around 10−9 s/s/y for mammals [Citation165,Citation168]. However the evolutionary reconstruction of the NIRVs remains tricky. EVEs are considered as « fossil records » of ancient infections [Citation169]. Several different methods have been described to date EVEs [Citation110,Citation170]. The minimum insertion date of the EVE can be evaluated if the divergence time of the two species sharing the same taxonomic position is known [Citation110]. Studies on EVE evolution revealed that many viral families are more ancient than previously thought. As an example, the lentivirus family classified as retroviruses dated to a hundred years by molecular clock dating techniques [Citation171] appeared several million years ago since endogenous lentiviruses were discovered in the grey mouse lemur (Microcebus murinus) from Madagascar [Citation172]. This can be extended to other viruses: Hepadnaviridae [Citation114,Citation173] and Bornaviridae [Citation110].
Conclusion
Aedes albopictus and Ae. aegypti are two mosquito species that have different histories. They vector several major human arboviruses, including CHIKV and DENV, for which they exhibit different vector competence. Whereas both species highly transmit CHIKV, Ae. albopictus is considered as a less efficient vector for DENV [Citation21]. Along with environmental factors such as the temperature, epigenetic factors like the mosquito microbiota [Citation1,Citation66,Citation67], and genetic factors like Quantitative Trait Loci (QTLs) [Citation86] are important to determine the vector competence. More importantly, the recent discovery of NIRVS highlights their potential role as modulator of vector competence to arboviruses. It has been suggested that their association with retrotransposons allowed them to be reverse transcribed into viral DNA (vDNA) and then be integrated into mosquito genomes. Moreover, NIRVS were found to produce vsiRNAs and vpiRNAs, which are important molecules in the RNAi-based immunity in Aedes mosquitoes [Citation119,Citation120,Citation125]. In rare cases, NIRVS are translated into proteins that act as inhibitor of viral replication [Citation161]. However, some questions remain unsolved, such as to which aim NIRVS are involved in the antiviral immunity. NIRVS could act as a warning signal and prime the antiviral immunity for allowing the host to control viral replication before the infection becomes deleterious and harmful for the vector host. It reminds us the adaptive immunity mechanisms such as CRISPR-Cas systems in prokaryotic cells. Alternatively, NIRVS could also act as a keeper of persistent infection by maintaining a low level of viral replication, diminishing the negative impacts on the mosquito fitness. Nevertheless, analysis of natural mosquito populations revealed a high diversity of NIRVS at the intra- and inter-population levels (Houé et al. unpublished data; [Citation174]), suggesting many DNA recombination in NIRVS-surrounding areas. While many NIRVS have been found homologous to insect-specific flavi- and rhabdovirus [Citation119], which are genetically related to pathogenic viruses, none was found homologous to Togaviridae family that contains only two insect-specific viruses described so far [Citation175,Citation176]. This could explain why CHIKV is highly transmitted by both Ae. aegypti and Ae. albopictus, compared to the Flaviviridae family that harbours many insect-specific viruses [Citation177].
Supplemental Material
Download MS Word (11.9 KB)Acknowledgments
The authors thank Pei-Shi Yen and Xavier de Lamballerie for critical discussion.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Zhang G, Hussain M, O'Neill SL, et al. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci U S A. 2013;110:10276–10281. doi:10.1073/pnas.1303603110.
- Leparc-Goffart I, Nougairede A, Cassadou S, et al. Chikungunya in the Americas. Lancet. 2014;383:514. doi:10.1016/S0140-6736(14)60185-9.
- Solignat M, Gay B, Higgs S, et al. Replication cycle of chikungunya: a re-emerging arbovirus. Virology. 2009;393:183–197. doi:10.1016/j.virol.2009.07.024.
- Gardner CL, Ryman KD. Yellow fever: a reemerging threat. Clin Lab Med. 2010;30:237–260. doi:10.1016/j.cll.2010.01.001.
- Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi:10.1038/nature12060.
- Simmons CP, Farrar JJ, Nguyen v V, et al. Dengue. N. Engl. J. Med. 2012;366:1423–1432. doi:10.1056/NEJMra1110265.
- Diniz DFA, de Albuquerque CMR, Oliva LO, et al. Diapause and quiescence: dormancy mechanisms that contribute to the geographical expansion of mosquitoes and their evolutionary success. Parasit Vectors. 2017;10:310. doi:10.1186/s13071-017-2235-0.
- Rezende GL, Martins AJ, Gentile C, et al. Embryonic desiccation resistance in Aedes aegypti: presumptive role of the chitinized serosal cuticle. BMC Dev Biol. 2008;8:82. doi:10.1186/1471-213X-8-82.
- Sota T, Mogi M. Interspecific variation in desiccation survival time of Aedes (Stegomyia) mosquito eggs is correlated with habitat and egg size. Oecologia. 1992;90:353–358. doi:10.1007/BF00317691.
- Urbanski JM, Benoit JB, Michaud MR, et al. The molecular physiology of increased egg desiccation resistance during diapause in the invasive mosquito, Aedes albopictus. Proc Biol Sci. 2010;277:2683–2692. doi:10.1098/rspb.2010.0362.
- Kraemer MU, Sinka ME, Duda KA, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife. 2015;4:e08347. doi:10.7554/eLife.08347.
- Christophers SR. Aedes aegypti (L.) the yellow fever mosquito: its life history, bionomics and structure. London (UK): Cambridge University Press; 1960; p. 750.
- Murphy EJ. History of African Civilization. New York: Crowell; 1972.
- Powell JR, Gloria-Soria A, Kotsakiozi P. Recent history of Aedes aegypti: vector Genomics and Epidemiology records. Bioscience. 2018;68:854–860. doi:10.1093/biosci/biy119.
- Smith CE. The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes aegypti. J Trop Med Hyg. 1956;59:243–251.
- Mattingly PF. New records and a new species of the subgenus Stegomyia (Diptera, Culicidae) from the Ethiopian region. Ann Trop Med Parasitol. 1953;47:294–298.
- Metselaar D, Grainger CR, Oei KG, et al. An outbreak of type 2 dengue fever in the Seychelles, probably transmitted by Aedes albopictus (Skuse). Bull. World Health Organ. 1980;58:937–943.
- Elliott SA. Aedes albopictus in the Solomon and Santa Cruz islands, South Pacific. Trans. R. Soc. Trop. Med. Hyg.. 1980;74:747–748.
- Caminade C, Medlock JM, Ducheyne E, et al. Suitability of European climate for the Asian tiger mosquito Aedes albopictus: recent trends and future scenarios. J R Soc Interface. 2012;9:2708–2717. doi:10.1098/rsif.2012.0138.
- Benedict MQ, Levine RS, Hawley WA, et al. Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus. Vector Borne Zoonotic Dis. 2007;7:76–85. doi:10.1089/vbz.2006.0562.
- Paupy C, Delatte H, Bagny L, et al. Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes Infect. 2009;11:1177–1185. doi:10.1016/j.micinf.2009.05.005.
- Cancrini G, Romi R, Gabrielli S, et al. First finding of Dirofilaria repens in a natural population of Aedes albopictus. Med Vet Entomol. 2003;17:448–451.
- Pietrobelli M. Importance of Aedes albopictus in veterinary medicine. Parassitologia. 2008;50:113–115.
- Christie J. On epidemics of dengue Fever; their Diffusion and Etiology. Ind Med Gaz. 1882;17:76–79.
- Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol. 2007;88:2363–2377. doi:10.1099/vir.0.82858-0.
- Kittisriworapod S, Thammapalo S. Fever with rash outbreak investigation report, Chumpare District, Khon Kaen Province. (1991).
- Muyembe-Tamfum JJ, Peyrefitte CN, Yogolelo R, et al. [Epidemic of chikungunya virus in 1999 and 200 in the Democratic Republic of the Congo]. Med Trop (Mars). 2003;63:637–638.
- Sergon K, Njuguna C, Kalani R, et al. Seroprevalence of chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am J Trop Med Hyg. 2008;78:333–337.
- Sergon K, Yahaya AA, Brown J, et al. Seroprevalence of chikungunya virus infection on Grande Comore Island, union of the Comoros, 2005. Am J Trop Med Hyg. 2007;76:1189–1193.
- Sang RC, Ahmed O, Faye O, et al. Entomologic investigations of a chikungunya virus epidemic in the union of the Comoros, 2005. Am J Trop Med Hyg. 2008;78:77–82.
- Singh KR, Pavri KM. Experimental studies with chikungunya virus in Aedes aegypti and Aedes albopictus. Acta Virol. 1967;11:517–526.
- Delatte H, Paupy C, Dehecq JS, et al. [Aedes albopictus, vector of chikungunya and dengue viruses in Reunion Island: biology and control]. Parasite. 2008;15:3–13. doi:10.1051/parasite/2008151003.
- Borgherini G, Poubeau P, Staikowsky F, et al. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin Infect Dis. 2007;44:1401–1407. doi:10.1086/517537.
- Bagny Beilhe L, Delatte H, Juliano SA, et al. Ecological interactions in Aedes species on Reunion Island. Med Vet Entomol. 2013;27:387–397. doi:10.1111/j.1365-2915.2012.01062.x.
- Schuffenecker I, Iteman I, Michault A, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3:e263. doi:10.1371/journal.pmed.0030263.
- Tsetsarkin KA, Vanlandingham DL, McGee CE, et al. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi:10.1371/journal.ppat.0030201.
- Vazeille M, Moutailler S, Coudrier D, et al. Two chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One. 2007;2:e1168. doi:10.1371/journal.pone.0001168.
- Powers AM. Risks to the Americas associated with the continued expansion of chikungunya virus. J Gen Virol. 2015;96:1–5. doi:10.1099/vir.0.070136-0.
- Gubler DJ. The global pandemic of dengue/dengue haemorrhagic fever: current status and prospects for the future. Ann Acad Med Singapore. 1998;27:227–234.
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496.
- Hotta S. Dengue vector mosquitoes in Japan: the role of Aedes albopictus and Aedes aegypti in the 1942–1944 dengue epidemics of Japan main islands. Med Entomol Zool. 1998;49:267–274.
- Luo L, Jiang L-Y, Xiao X-C, et al. The dengue preface to endemic in mainland China: the historical largest outbreak by Aedes albopictus in Guangzhou, 2014. Infect Dis Poverty. 2017;6:148. doi:10.1186/s40249-017-0352-9.
- Lambrechts L, Scott TW, Gubler DJ. Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl Trop Dis. 2010;4:e646. doi:10.1371/journal.pntd.0000646.
- La Ruche G, Souarès Y, Armengaud A, et al. First two autochthonous dengue virus infections in metropolitan France, September 2010. Euro Surveillance : Bulletin Europeen sur les Maladies Transmissibles = European Communicable Disease Bulletin. 2010;15:19676.
- Gjenero-Margan I, Aleraj B, Krajcar D, et al. Autochthonous dengue fever in Croatia, August-September 2010. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2011;16(9):pii: 19805.
- Rezza G. Aedes albopictus and the reemergence of dengue. BMC Public Health. 2012;12:72. doi:10.1186/1471-2458-12-72.
- Hardy JL. In: Monath TP, editor. The arboviruses: Epidemiology and Ecology. Boca Raton (FL): CRC Press; 1988. Vol. 2, p. 87–126.
- Woodring JL, Higgs S, Beaty BJ. In: Marquardt WC, Beaty BJ, editors. The Biology of Disease vectors. Niwot: University Press of Colorado; 1996; p. 632.
- Lambrechts L, Chevillon C, Albright RG, et al. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol Biol. 2009;9:160. doi:10.1186/1471-2148-9-160.
- Zouache K, Fontaine A, Vega-Rúa A, et al. Three-way interactions between mosquito population, viral strain and temperature underlying chikungunya virus transmission potential. Proc Biol Sci / R Soc. 2014;281. doi:10.1098/rspb.2014.1078.
- Coffey LL, Failloux AB, Weaver SC. Chikungunya virus-vector interactions. Viruses. 2014;6:4628–4663. doi:10.3390/v6114628.
- Sunarto J, Gubler DJ, Nalim S, et al. Epidemic dengue hemorrhagic fever in rural Indonesia. III. Entomological studies. Am J Trop Med Hyg 1979;28(4):717–724.
- Rosen L, Roseboom LE, Gubler DJ, et al. Comparative susceptibility of mosquito species and strains to oral and parenteral infection with dengue and Japanese encephalitis viruses. Am J Trop Med Hyg. 1985;34:603–615.
- Alto BW, Reiskind MH, Lounibos LP. Size alters susceptibility of vectors to dengue virus infection and dissemination. Am J Trop Med Hyg. 2008;79:688–695.
- Vazeille M, Rosen L, Mousson L, et al. Low oral receptivity for dengue type 2 viruses of Aedes albopictus from Southeast Asia compared with that of Aedes aegypti. Am J Trop Med Hyg. 2003;68:203–208.
- Whitehead RH, Yuill TM, Gould DJ, et al. Experimental infection of Aedes aegypti and Aedes albopictus with dengue viruses. T Roy Soc Trop Med Hyg. 1971;65:661–667.
- Lindh JM, Borg-Karlson AK, Faye I. Transstadial and horizontal transfer of bacteria within a colony of Anopheles gambiae (Diptera: Culicidae) and oviposition response to bacteria-containing water. Acta Trop. 2008;107:242–250. doi:10.1016/j.actatropica.2008.06.008.
- Alvarez-Perez S, Herrera CM, de Vega C. Zooming-in on floral nectar: a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol Ecol. 2012;80:591–602. doi:10.1111/j.1574-6941.2012.01329.x.
- Douglas AE. Lessons from studying insect symbioses. Cell Host Microbe. 2011:359–367. doi:10.1016/j.chom.2011.09.001.
- Douglas AE. The molecular basis of bacterial-insect symbiosis. J Mol Biol. 2014;426:3830–3837. doi:10.1016/j.jmb.2014.04.005.
- Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi:10.1146/annurev.ento.49.061802.123416.
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi:10.1146/annurev.genet.41.110306.130119.
- Moya A, Pereto J, Gil R, et al. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi:10.1038/nrg2319.
- Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435.
- Charroux B, Royet J. Drosophila immune response: from systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly (Austin). 2010;4:40–47.
- Pan X, Zhou G, Wu J, et al. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2012;109:E23–E31. doi:10.1073/pnas.1116932108.
- Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi:10.1371/journal.ppat.1000098.
- Fragkoudis R, Attarzadeh-Yazdi G, Nash AA, et al. Advances in dissecting mosquito innate immune responses to arbovirus infection. J Gen Virol. 2009;90:2061–2072. doi:10.1099/vir.0.013201-0.
- Avadhanula V, Weasner BP, Hardy GG, et al. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5:e1000582. doi:10.1371/journal.ppat.1000582.
- Costa A, Jan E, Sarnow P, et al. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One. 2009;4:e7436. doi:10.1371/journal.pone.0007436.
- Paradkar PN, Trinidad L, Voysey R, et al. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc Natl Acad Sci U S A. 2012;109:18915–18920. doi:10.1073/pnas.1205231109.
- Zambon RA, Nandakumar M, Vakharia VN, et al. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A. 2005;102:7257–7262. doi:10.1073/pnas.0409181102.
- Sim S, Jupatanakul N, Dimopoulos G. Mosquito immunity against arboviruses. Viruses. 2014;6:4479–4504. doi:10.3390/v6114479.
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi:10.1242/dev.02411.
- Hombria JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569–R575.
- Zhu F, Ding H, Zhu B. Transcriptional profiling of Drosophila S2 cells in early response to Drosophila C virus. Virol J. 2013;10:210. doi:10.1186/1743-422X-10-210.
- Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci U S A. 2009;106:17841–17846. doi:10.1073/pnas.0905006106.
- Huang Z, Kingsolver MB, Avadhanula V, et al. An antiviral role for antimicrobial peptides during the arthropod response to alphavirus replication. J Virol. 2013;87:4272–4280. doi:10.1128/JVI.03360-12.
- McFarlane M, Arias-Goeta C, Martin E, et al. Characterization of Aedes aegypti innate-immune pathways that limit chikungunya virus replication. PLoS Negl Trop Dis. 2014;8:e2994. doi:10.1371/journal.pntd.0002994.
- Fragkoudis R, Chi Y, Siu RW, et al. Semliki Forest virus strongly reduces mosquito host defence signaling. Insect Mol Biol. 2008;17:647–656. doi:10.1111/j.1365-2583.2008.00834.x.
- Barletta AB, Nascimento-Silva MC, Talyuli OA, et al. Microbiota activates IMD pathway and limits Sindbis infection in Aedes aegypti. Parasit Vectors. 2017;10:103. doi:10.1186/s13071-017-2040-9.
- Sanders HR, Foy BD, Evans AM, et al. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochem Molec. 2005;35:1293–1307. doi:10.1016/j.ibmb.2005.07.006.
- Failloux AB, Vazeille M, Rodhain F. Geographic genetic variation in populations of the dengue virus vector Aedes aegypti. J Mol Evol. 2002; 55(6):653–663.
- Bosio CF, Fulton RE, Salasek ML, et al. Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti. Genetics. 2000;156:687–698.
- Gomez-Machorro C, Bennett KE, del Lourdes Munoz M, et al. Quantitative trait loci affecting dengue midgut infection barriers in an advanced intercross line of Aedes aegypti. Insect Mol Biol. 2004;13:637–648. doi:10.1111/j.0962-1075.2004.00522.x.
- Bennett KE, Flick D, Fleming KH, et al. Quantitative trait loci that control dengue-2 virus dissemination in the mosquito Aedes aegypti. Genetics. 2005;170:185–194. doi:10.1534/genetics.104.035634.
- Nene V, Wortman JR, Lawson D, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science (New York, N.Y.). 2007;316:1718–1723. doi:10.1126/science.1138878.
- Dritsou V, Topalis P, Windbichler N, et al. A draft genome sequence of an invasive mosquito: an Italian Aedes albopictus. Pathog Glob Health. 2015;109:207–220. doi:10.1179/2047773215Y.0000000031.
- Chen XG, Jiang X, Gu J, et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc Natl Acad Sci U S A. 2015;112:E5907–E5915. doi:10.1073/pnas.1516410112.
- Holt RA, Subramanian GM, Halpern A, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science (New York, N.Y.). 2002;298:129–149. doi:10.1126/science.1076181.
- Arensburger P, Megy K, Waterhouse RM, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science (New York, N.Y.). 2010;330:86–88. doi:10.1126/science.1191864.
- McLain DK, Rai KS, Fraser MJ. Intraspecific and interspecific variation in the sequence and abundance of highly repeated DNA among mosquitoes of the Aedes albopictus subgroup. Heredity (Edinb). 1987;58(Pt 3):373–381.
- Black WCt, Ferrari JA, Rai KS, et al. Breeding structure of a colonising species: Aedes albopictus (Skuse) in the United States. Heredity (Edinb). 1988;60(Pt 2):173–181.
- McClintock B. Intranuclear systems controlling gene action and mutation. Brookhaven Symp Biol. 1956;(8):58–74.
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284(5757):601–603.
- Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607.
- Klaer R, Kuhn S, Tillmann E, et al. The sequence of IS4. Mol Gen Genet. 1981;181:169–175.
- Au TK, Agrawal P, Harshey RM. Chromosomal integration mechanism of infecting mu virion DNA. J Bacteriol. 2006;188:1829–1834. doi:10.1128/JB.188.5.1829-1834.2006.
- Bukhari A, Zipser D. Random insertion of Mu-1 DNA within a single gene. Nature (London). 1972;236:240–243.
- Finnegan DJ. Retrotransposons. Curr Biol. 2012;22:R432–R437. doi:10.1016/j.cub.2012.04.025.
- Craig NL, Craigie R, Gellert M, et al. Mobile DNA II. (American Society for Microbiology Press, DC Washington, 2002).
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:8714–8719. doi:10.1073/pnas.151269298.
- Pritham EJ, Putliwala T, Feschotte C. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene. 2007;390:3–17. doi:10.1016/j.gene.2006.08.008.
- Kidwell MG. Horizontal transfer. Curr Opin Genet Dev. 1992;2:868–873.
- Konkel MK, Batzer MA. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 2010;20:211–221. doi:10.1016/j.semcancer.2010.03.001.
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi:10.1038/nrg798.
- Rao PN, Rai K. Inter and intraspecific variation in nuclear DNA content in Aedes mosquitoes. Heredity (Edinb). 1987;59(Pt 2):253–258.
- Goubert C, Modolo L, Vieira C, et al. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Biol Evol. 2015;7:1192–1205. doi:10.1093/gbe/evv050.
- Biryukova I, Ye T. Endogenous siRNAs and piRNAs derived from transposable elements and genes in the malaria vector mosquito Anopheles gambiae. BMC Genomics. 2015;16:278. doi:10.1186/s12864-015-1436-1.
- Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi:10.1371/journal.pgen.1001191.
- Siomi MC, Sato K, Pezic D, et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi:10.1038/nrm3089.
- Bock M, Stoye JP. Endogenous retroviruses and the human germline. Curr Opin Genet Dev. 2000;10:651–655.
- Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi:10.1371/journal.ppat.1001030.
- Gilbert C, Feschotte C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010;8. doi:10.1371/journal.pbio.1000495.
- Crochu S, Cook S, Attoui H, et al. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 2004;85:1971–1980. doi:10.1099/vir.0.79850-0.
- Suzuki Y, Frangeul L, Dickson LB, et al. Uncovering the Repertoire of endogenous flaviviral elements in Aedes mosquito genomes. J Virol. 2017;91. doi:10.1128/JVI.00571-17.
- Bolling BG, Weaver SC, Tesh RB, et al. Insect-Specific Virus discovery: significance for the arbovirus Community. Viruses. 2015;7:4911–4928. doi:10.3390/v7092851.
- Vasilakis N, Tesh RB. Insect-specific viruses and their potential impact on arbovirus transmission. Curr Opin Virol. 2015;15:69–74. doi:10.1016/j.coviro.2015.08.007.
- Palatini U, Miesen P, Carballar-Lejarazu R, et al. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics. 2017;18:512. doi:10.1186/s12864-017-3903-3.
- Whitfield ZJ, Dolan PT, Kunitomi M, et al. The diversity, structure, and function of Heritable adaptive immunity sequences in the Aedes aegypti genome. Curr Biol. 2017;27:3511–3519 e3517. doi:10.1016/j.cub.2017.09.067.
- Holmes EC. The evolution of endogenous viral elements. Cell Host Microbe. 2011;10:368–377. doi:10.1016/j.chom.2011.09.002.
- Fort P, Albertini A, Van-Hua A, et al. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution, and potential functionality. Mol Biol Evol. 2012;29:381–390. doi:10.1093/molbev/msr226.
- Conzelmann KK. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genet. 1998;32:123–162. doi:10.1146/annurev.genet.32.1.123.
- Brackney DE, Scott JC, Sagawa F, et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4:e856. doi:10.1371/journal.pntd.0000856.
- Goic B, Stapleford KA, Frangeul L, et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat Commun. 2016;7:12410. doi:10.1038/ncomms12410.
- Nag DK, Brecher M, Kramer LD. DNA forms of arboviral RNA genomes are generated following infection in mosquito cell cultures. Virology. 2016;498:164–171. doi:10.1016/j.virol.2016.08.022.
- Nag DK, Kramer LD. Patchy DNA forms of the zika virus RNA genome are generated following infection in mosquito cell cultures and in mosquitoes. J Gen Virol. 2017;98:2731–2737. doi:10.1099/jgv.0.000945.
- Geuking MB, Weber J, Dewannieux M, et al. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science (New York, N.Y.). 2009;323:393–396. doi:10.1126/science.1167375.
- Zhdanov VM. Integration of viral genomes. Nature. 1975;256:471–473.
- Goic B, Vodovar N, Mondotte JA, et al. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol. 2013;14:396–403. doi:10.1038/ni.2542.
- Bill CA, Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A. 2004;101:11135–11140. doi:10.1073/pnas.0403925101.
- Kotin RM, Linden RM, Berns KI. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. Embo J. 1992;11:5071–5078.
- Urcelay E, Ward P, Wiener SM, et al. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046.
- Young SM Jr., Samulski RJ. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc Natl Acad Sci U S A 2001;98:13525–13530. doi:10.1073/pnas.241508998.
- Morissette G, Flamand L. Herpesviruses and chromosomal integration. J Virol. 2010;84:12100–12109. doi:10.1128/JVI.01169-10.
- Klenerman P, Hengartner H, Zinkernagel RM. A non-retroviral RNA virus persists in DNA form. Nature. 1997;390:298–301. doi:10.1038/36876.
- Maori E, Tanne E, Sela I. Reciprocal sequence exchange between non-retro viruses and hosts leading to the appearance of new host phenotypes. Virology. 2007;362:342–349. doi:10.1016/j.virol.2006.11.038.
- Bronkhorst AW, van Rij RP. The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol. 2014;7:19–28. doi:10.1016/j.coviro.2014.03.010.
- Olson KE, Blair CD. Arbovirus-mosquito interactions: RNAi pathway. Curr Opin Virol. 2015;15:119–126. doi:10.1016/j.coviro.2015.10.001.
- Obbard DJ, Gordon KH, Buck AH, et al. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115. doi:10.1098/rstb.2008.0168.
- Sarot E, Payen-Groschene G, Bucheton A, et al. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321.
- Saito K, Nishida KM, Mori T, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi:10.1101/gad.1454806.
- Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi:10.1016/j.cell.2007.01.043.
- Vagin VV, Sigova A, Li C, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science (New York, N.Y.). 2006;313:320–324. doi:10.1126/science.1129333.
- Pelisson A, Sarot E, Payen-Groschene G, et al. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi:10.1128/JVI.01980-06.
- Akbari OS, Antoshechkin I, Amrhein H, et al. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda). 2013;3:1493–1509. doi:10.1534/g3.113.006742.
- Arensburger P, Hice RH, Wright JA, et al. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genomics. 2011;12:606. doi:10.1186/1471-2164-12-606.
- Morazzani EM, Wiley MR, Murreddu MG, et al. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012;8:e1002470. doi:10.1371/journal.ppat.1002470.
- Schnettler E, Donald CL, Human S, et al. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J Gen Virol. 2013;94:1680–1689. doi:10.1099/vir.0.053850-0.
- Vodovar N, Bronkhorst AW, van Cleef KW, et al. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One. 2012;7:e30861. doi:10.1371/journal.pone.0030861.
- Hess AM, Prasad AN, Ptitsyn A, et al. Small RNA profiling of dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011;11:45. doi:10.1186/1471-2180-11-45.
- Leger P, Lara E, Jagla B, et al. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J Virol. 2013;87:1631–1648. doi:10.1128/JVI.02795-12.
- Miesen P, Joosten J, van Rij RP. PIWIs Go viral: arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog. 2016;12:e1006017. doi:10.1371/journal.ppat.1006017.
- Miesen P, Ivens A, Buck AH, et al. Small RNA Profiling in dengue Virus 2-infected Aedes mosquito cells Reveals viral piRNAs and Novel host miRNAs. PLoS Negl Trop Dis. 2016;10:e0004452. doi:10.1371/journal.pntd.0004452.
- Petit M, Mongelli V, Frangeul L, et al. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2016;113:E4218–E4227. doi:10.1073/pnas.1607952113.
- Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011;6:265–277. doi:10.2217/fmb.11.11.
- Kingsolver MB, Huang Z, Hardy RW. Insect antiviral innate immunity: pathways, effectors, and connections. J Mol Biol. 2013;425:4921–4936. doi:10.1016/j.jmb.2013.10.006.
- van Cleef KW, van Mierlo JT, van den Beek M, et al. Identification of viral suppressors of RNAi by a reporter assay in Drosophila S2 cell culture. Methods Mol Biol. 2011;721:201–213. doi:10.1007/978-1-61779-037-9_12.
- Aswad A, Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol Evol. 2012;27:627–636. doi:10.1016/j.tree.2012.07.007.
- Taylor GM, Gao Y, Sanders DA. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J Virol. 2001;75:11244–11248. doi:10.1128/JVI.75.22.11244-11248.2001.
- Fujino K, Horie M, Honda T, et al. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc Natl Acad Sci U S A. 2014;111:13175–13180. doi:10.1073/pnas.1407046111.
- Mura M, Murcia P, Caporale M, et al. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A. 2004;101:11117–11122. doi:10.1073/pnas.0402877101.
- Lequime S, Lambrechts L. Discovery of flavivirus-derived endogenous viral elements in Anopheles mosquito genomes supports the existence of Anopheles-associated insect-specific flaviviruses. Virus Evol 2017;3, vew035. doi:10.1093/ve/vew035.
- Hanada K, Suzuki Y, Gojobori T. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol Biol Evol. 2004;21:1074–1080. doi:10.1093/molbev/msh109.
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi:10.1038/nrg2323.
- Jenkins GM, Rambaut A, Pybus OG, et al. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi:10.1007/s00239-001-0064-3.
- Sanjuan R. From molecular genetics to phylodynamics: evolutionary relevance of mutation rates across viruses. PLoS Pathog. 2012;8:e1002685. doi:10.1371/journal.ppat.1002685.
- Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci U S A. 2002;99:803–808. doi:10.1073/pnas.022629899.
- Emerman M, Malik HS. Paleovirology–modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi:10.1371/journal.pbio.1000301.
- Aiewsakun P, Katzourakis A. Endogenous viruses: Connecting recent and ancient viral evolution. Virology. 2015;479–480:26–37. doi:10.1016/j.virol.2015.02.011.
- Wertheim JO, Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol. 2009;5:e1000377. doi:10.1371/journal.pcbi.1000377.
- Gifford RJ, Katzourakis A, Tristem M, et al. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci U S A. 2008;105:20362–20367. doi:10.1073/pnas.0807873105.
- Zhou Y, Holmes EC. Bayesian estimates of the evolutionary rate and age of hepatitis B virus. J Mol Evol. 2007;65:197–205. doi:10.1007/s00239-007-0054-1.
- Pischedda E. Scolari F, Valerio F, et al. Evolutionary landscape of mosquito viral integrations. BioRxiv. 2018. doi:10.1101/385666.
- Hermanns K, Zirkel F, Kopp A, et al. Discovery of a novel alphavirus related to Eilat virus. J Gen Virol. 2017;98:43–49. doi:10.1099/jgv.0.000694.
- Nasar F, Palacios G, Gorchakov RV, et al. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci U S A. 2012; 109:14622–14627. doi:10.1073/pnas.1204787109.
- Blitvich BJ, Firth AE. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses. 2015;7:1927–1959. doi:10.3390/v7041927.