ABSTRACT
Clostridium difficile ribotype (RT) 017 is an important toxigenic C. difficile RT which, due to a deletion in the repetitive region of the tcdA gene, only produces functional toxin B. Strains belonging to this RT were initially dismissed as nonpathogenic and circulated largely undetected for almost two decades until they rose to prominence following a series of outbreaks in the early 2000s. Despite lacking a functional toxin A, C. difficile RT 017 strains have been shown subsequently to be capable of causing disease as severe as that caused by strains producing both toxins A and B. While C. difficile RT 017 strains can be found in almost every continent today, epidemiological studies suggest that the RT is endemic in Asia and that the global spread of this MLST clade 4 lineage member is a relatively recent event. C. difficile RT 017 transmission appears to be mostly from human to human with only a handful of reports of isolations from animals. An important feature of C. difficile RT 017 strains is their resistance to several antimicrobials and this has been documented as a possible factor driving multiple outbreaks in different parts of the world. This review summarizes what is currently known regarding the emergence and evolution of strains belonging to C. difficile RT 017 as well as features that have allowed it to become an RT of global importance.
Introduction
Clostridium difficile is an important cause of antimicrobial-associated diarrhoea (AAD) in both humans and animals [Citation1]. In humans, the disease can progress from watery diarrhoea to life-threatening pseudomembranous colitis (PMC) and toxic megacolon [Citation2]. C. difficile infection (CDI) is a toxin-mediated disease and major virulence factors include toxin A (TcdA, 308 kDa) and toxin B (TcdB, 270 kDa) [Citation3]. An additional binary toxin (C. difficile transferase, CDT) is produced by some strains only. CDT-producing strains of C. difficile account for an increasing proportion of human infections in some parts of the world (currently ca. 20% of CDI cases in non-outbreak situations) but are common in animals [Citation4,Citation5]. C. difficile can be classified into different PCR ribotypes (RTs) using banding patterns of the amplified intergenic spacer region between the 16S and 23S rRNA genes [Citation6]. Currently, over 600 RTs exist in the United Kingdom-based C. difficile Ribotyping Network (CDRN) database [Citation7].
C. difficile RT 017 ranks among the most successful RTs of C. difficile. A toxigenic strain that produces only TcdB [Citation8], RT 017 causes disease as severe as other toxigenic strains [Citation9–12]. Although C. difficile RT 017 appears to have originated in Asia, it has spread globally and been responsible for multiple outbreaks around the world [Citation13–23]. Few studies have been conducted to identify factors that may have contributed to the success of RT 017 [Citation16,Citation18]. This review summarizes what is known about C. difficile RT 017 regarding its history, characteristics, evolution, emergence and global dissemination.
Brief history of C. difficile infection and the emergence of C. difficile RT 017
C. difficile (then named Bacillus difficilis) was first described in 1935 as part of neonatal gut flora. It produced a potent cytotoxin that caused tissue oedema, convolution and death when injected subcutaneously into guinea pigs and rabbits [Citation24]. However, there were no reports of human gastrointestinal infections associated with C. difficile until 1978 when, after a period of intense trans-Atlantic competition between researchers, C. difficile was identified in faecal specimens from patients with PMC [Citation25].
Not all strains of C. difficile produce toxins and cause disease. Initially, it was thought that all toxigenic strains of C. difficile produced both major toxins [Citation26]. For two decades after the association between C. difficile and PMC was shown, it was believed that TcdA was required to cause initial damage to the intestinal mucosa before TcdB could exert its potent cytotoxic effect [Citation27], and the significance of TcdA-negative, TcdB-positive (A-B+) stains was not apparent [Citation17]. To further support this belief, the first few strains of C. difficile isolated with an A-B+ phenotype were associated only with asymptomatic carriage [Citation28]. During this same period, there was a move away from using the faecal TcdB cytotoxicity assay and/or culture of C. difficile for diagnostic purposes due to the time and expense involved in maintaining and using cell lines, and the long turnaround time of culture. Concomitantly, there was an emphasis on developing rapid immunoassays for the detection of TcdA [Citation29]. TcdA was chosen because of the continued mistaken belief that C. difficile produced either both TcdA and TcdB, or no toxins, because it was easier to manufacture antibodies against TcdA, and because detection of TcdA had greater sensitivity compared to detection of TcdB [Citation30]. These tools made the detection of C. difficile easier, but with far less overall sensitivity, and further obscured the significance of A-B+ C. difficile strains.
The importance of A-B+ strains of C. difficile was finally appreciated at the end of the twentieth century when 16 patients in a Canadian tertiary-care hospital developed PMC with an A-B+ strain. Stool samples from these patients tested negative for C. difficile TcdA but were later shown via a cytotoxin assay to contain C. difficile that produced a functional TcdB only [Citation17]. Similar findings were published from other countries [Citation13,Citation16] and further studies confirmed these strains as A-B+ C. difficile RT 017 [Citation8]. At the same time, a study reported that not only could TcdB exert its cytotoxic effect in the absence of TcdA, but also that human intestinal mucosa was around 10 times more sensitive to TcdB than TcdA [Citation31]. This was the first time that the clinical significance of A-B+ C. difficile became evident [Citation32]. Over the last 20 years, C. difficile RT 017 has been isolated from many parts of the world, however, it is likely that C. difficile RT 017 originated from a single geographical region and its global dispersal has been a relatively recent event [Citation33].
Characteristics of C. difficile RT 017
Epidemiological typing of C. difficile RT 017
Currently, PCR ribotyping is a method of typing C. difficile that is widely used in many parts of the world due to its relative simplicity and high discriminatory power [Citation34]. However, ribotyping requires comparison of banding patterns with those of standard strains present in a library of patterns that was established in 1999 [Citation6]. Thus, reports of C. difficile before or around that time classified C. difficile by various other methods [Citation17,Citation35]. summarizes these different methods used when referring to C. difficile RT 017. Early ribotyping studies in Japan used their own nomenclature and assigned “fr” to RT 017 [Citation36].
Table 1. C. difficile RT 017 categorized by other classification methods.
Before genotype-based methods, C. difficile was classified using phenotypic methods that, in general, had poor reproducibility, low typeability, and lacked sufficient discriminatory power to be applied to epidemiological studies [Citation42]. However, serogrouping was widely used early and showed a good correlation with toxigenicity [Citation43]. Serogrouping classified C. difficile RT 017 as either serogroup F or X [Citation37].
Many genotypic methods, including ribotyping, use unique banding patterns of different PCR products to classify C. difficile strains. Toxinotyping detects differences in the Pathogenicity Locus (PaLoc) and classifies C. difficile RT 017 as toxinotype VIII [Citation38]. Pulsed-field gel electrophoresis is more commonly used in North America and classifies C. difficile RT 017 as North American pulsed-field gel electrophoresis type 9 (NAP 9) [Citation39]. Restriction endonuclease analysis (REA) typing has greater discriminatory power than ribotyping and divides C. difficile RT 017 into several REA types which are grouped as REA groups CF and CG [Citation37].
Multi-locus sequence typing (MLST) is another genotype-based method involving 7 housekeeping genes. However, it is not based on banding patterns but rather the unique sequences of these genes and thus has been used mainly in evolutionary studies. This method classifies C. difficile RT 017 as sequence type (ST) 37 belonging to evolutionary clade 4 [Citation40]. MLST has good discriminatory power, however, it is relatively more complicated to perform [Citation34]. The advent of next-generation sequencing makes in silico MLST now more accessible [Citation44].
A recent study in China reported that RT 017 can also be identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) with high sensitivity and specificity [Citation45]. However, this study did not include other C. difficile strains from clade 4 and another Chinese study suggested that different clade 4 strains may not be distinguishable by this method [Citation46].
C. difficile RT 017 toxin
C. difficile RT 017 is classified as A-B+ C. difficile as it produces only a functional TcdB [Citation8]. Its TcdB also gives a different cytopathic effect (CPE) in cell cytotoxin assays using various cell lines compared to other strains that is often referred to as a variant CPE [Citation16,Citation47]. Studies on the tcdA gene of C. difficile RT 017 revealed a 1.8 kb deletion in the repeating region (3′ end) () and a point mutation in the 5′ end which results in a premature stop codon [Citation49,Citation50]. The 1.8 kb deletion corresponds to a deletion of the carboxy repetitive oligopeptide (CROP) region of TcdA, which is the recognition site of many TcdA enzyme immunoassays (EIAs), making the toxin undetectable by these EIAs [Citation47]. The nonsense mutation at 5′ end corresponds with a loss of catalytic action of the TcdA, thus making the toxin non-functional [Citation47,Citation49].
Figure 1. Comparative analysis of the PaLoc from C. difficile RT 017 and A + B + C. difficile strains. Arrows indicate open reading frames (ORFs) and the direction of transcription. The different enzymatic domain of the tcdB gene is responsible for the different CPE [Citation48]. The nonsense mutation near the 5′ terminal of the tcdA gene is responsible for the loss of function of TcdA [Citation49]. The 1.8 kb deletion near the 3′ terminal of the tcdA gene makes TcdA undetectable by many toxin EIAs [Citation47].
![Figure 1. Comparative analysis of the PaLoc from C. difficile RT 017 and A + B + C. difficile strains. Arrows indicate open reading frames (ORFs) and the direction of transcription. The different enzymatic domain of the tcdB gene is responsible for the different CPE [Citation48]. The nonsense mutation near the 5′ terminal of the tcdA gene is responsible for the loss of function of TcdA [Citation49]. The 1.8 kb deletion near the 3′ terminal of the tcdA gene makes TcdA undetectable by many toxin EIAs [Citation47].](/cms/asset/bfdcde05-1748-4342-90cc-6ee1230afcd0/temi_a_1621670_f0001_oc.jpg)
Notably, despite lacking a functional TcdA, most of the tcdA gene in C. difficile RT 017 remains intact and can be detected by PCR if primers specific to the non-repeating region of the tcdA gene are used. In such cases, C. difficile RT 017 could be incorrectly detected as both tcdA- and tcdB-positive C. difficile [Citation51]. While these primers are efficient for detection of toxigenic strains in clinical practice, the results may appear confusing in an epidemiological study. An additional primer set is needed to identify the deletion in the repeating region of tcdA gene and differentiate C. difficile RT 017 from true A+B+ C. difficile strains [Citation28,Citation52].
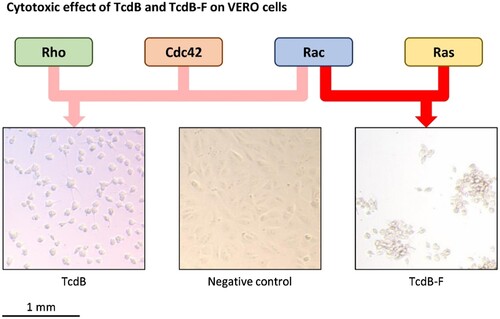
Interestingly, the TcdB of RT 017 (TcdB-F) is different from the TcdB commonly found in most C. difficile strains. TcdB-F behaves as a “functional hybrid,” combining characteristics of both TcdB and the Clostridium sordellii lethal toxin, TclS. While TcdB-F binds to the same cellular receptors as TcdB, the two proteins display differences in their target specificity, with TcdB primarily glucosylating Rho, Rac and Cdc42 targets, and TcdB-F glucosylating Rac and Ras targets () [Citation48]. The difference in cellular targets is thought to be responsible for the different CPE observed for the two toxins [Citation50].
Figure 2. The cytotoxic effect of TcdB and TcdB-F on VERO cells. VERO cells were treated with the supernatant of 72-hour-old cultures of C. difficile strain 2149 (RT 014/020 which produces TcdB), C. difficile strain 1470 (RT 017 which produce TcdB-F), and C. difficile ATCC 700057 (RT 038 which is non-toxigenic) and incubated at 37°C for 24 hours before inspection under a light microscope. TcdB glycosylates Rho, Rac, and Cdc42 targets resulting in arborization of cells while TcdB-F glycosylates Rac and Ras targets resulting in rounding of cells without arborization.
Infection due to C. difficile RT 017
Despite producing toxin B only, several studies suggest that C. difficile RT 017 causes clinical disease that is indistinguishable from that caused by other C. difficile RTs [Citation9,Citation12]. In addition, C. difficile RT 017 causes disease as severe as that caused by “hypervirulent” C. difficile RT 027 [Citation10]. In an outbreak setting, mortality due to C. difficile RT 017 can be as high as 37.5% [Citation47], but this high mortality rate may have been due to the exclusion of mild cases. There have been no clinical studies of C. difficile RT 017 infection in South East Asia, where there is a high prevalence of RT 017 [Citation41,Citation53,Citation54]. Given that CDI in this region was, in general, associated with low mortality and recurrence [Citation55], it will be interesting to see whether the less severe CDI in this region is specifically associated with C. difficile RT 017 or if there are other unknown protective factors in the population or region, such as a high prevalence of carriage of non-toxigenic strains, which may occupy the same niche and competitively exclude toxigenic strains from the gut [Citation53,Citation56,Citation57].
Evolution and transmission of C. difficile RT 017
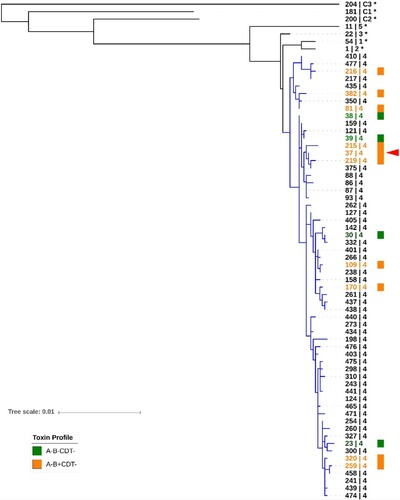
Based on MLST and Bayesian evolutionary model analysis (), C. difficile has evolved into at least five clades and three cryptic clades. This clade divergence occurred more than a million years ago [Citation34]. C. difficile RT 017 (ST 37; red arrowhead in ) is a member of C. difficile clade 4 along with many non-toxigenic, and some similar toxigenic, strains [Citation46,Citation58–61]. Despite limited data, it is clear that both A-B+CDT- and non-toxigenic strains of C. difficile (orange and green, respectively, in ) are equally distributed throughout clade 4, indicating that the clade 4 ancestor could either be a toxigenic (A-B+CDT-) or non-toxigenic strain. A recent study analyzed time-scaled core-genome phylogenies and suggested that the clade 4 ancestor was a non-toxigenic strain of C. difficile, and that acquisition of the PaLoc in C. difficile RT 017 occurred around 500 years ago [Citation59].
Figure 3. Sequence type diversity in evolutionary clade 4. Maximum-likelihood MLST phylogeny. Sequences were aligned using MUSCLE and tree was generated in MEGA7 with evolutionary distances calculated using the Tajima-Nei model. The scale shows the number of nucleotide substitution per site, based on concatenated MLST allele sequences (7 loci, 3501 bp). The tree is mid-point rooted and supported by 500 bootstrap replicates (only values >50 are shown). For global phylogenetic context, well-characterised representatives of MLST clade 1 (ST 54), 2 (ST 1), 3 (ST 22), 5 (ST 11), C1 (ST 181), C2 (ST 200), and C3 (ST 204) are also shown (*). Branches for clade 4 are shown in blue. Known toxin profiles of clade 4 strains are indicated by orange (A-B+CDT-) and green (A-B-CDT-) colour. RT 017 (ST 37) is indicated with a red arrowhead.
To date, the genomes of two C. difficile RT 017 strains (CF5, isolated in Belgium in 1995, and M68, isolated in Ireland in 2006) have been completely sequenced, providing important reference chromosomes for whole genome sequencing (WGS) studies of this lineage [Citation62]. shows the genome of C. difficile strain M68. Using WGS, Cairns et al. showed that 23 of 24 of C. difficile RT 017 strains from one hospital were closely related and formed a single cluster. The only unrelated C. difficile RT 017 strain was isolated from a patient with community-acquired CDI and this belonged to a cluster from outer London hospitals. These findings suggested that C. difficile RT 017 was mostly transmitted between patients in the same ward and between wards in the same hospital. The study further found that environmental contamination with clinical isolates was possible and that RT 017 could withstand decontamination with hydrogen peroxide vapour [Citation22].
Figure 4. A. Circular representation of the genome of C. difficile strain M68 (RT 017, ST 37, GenBank accession number NC017175.1). From outside to inside, the concentric circles represent (1) and (2) all coding sequences (CDS) transcribed in clockwise and counter-clockwise, (3) all rRNA, (4) all tRNA, (5) transposons (Tn6194 containing ermB gene represented in red and Tn6190 containing tetM gene represented in purple) and prophages (counterclockwise from top; ΦCDHM19 [58,163 bp, GC% = 31.34%], ΦCDHM13 [39,325 bp, GC% = 29.34%], and ΦMMP01 [55,106 bp, GC% = 28.87%]), and (6) GC content. B. Key characteristics of the genome.
![Figure 4. A. Circular representation of the genome of C. difficile strain M68 (RT 017, ST 37, GenBank accession number NC017175.1). From outside to inside, the concentric circles represent (1) and (2) all coding sequences (CDS) transcribed in clockwise and counter-clockwise, (3) all rRNA, (4) all tRNA, (5) transposons (Tn6194 containing ermB gene represented in red and Tn6190 containing tetM gene represented in purple) and prophages (counterclockwise from top; ΦCDHM19 [58,163 bp, GC% = 31.34%], ΦCDHM13 [39,325 bp, GC% = 29.34%], and ΦMMP01 [55,106 bp, GC% = 28.87%]), and (6) GC content. B. Key characteristics of the genome.](/cms/asset/506fc363-a219-4a2f-b0f1-93bdede3c8a4/temi_a_1621670_f0004_oc.jpg)
Another WGS study of 277 different C. difficile RT 017 strains isolated from around the world, including 24 from animals (cattle, dogs, and horses) showed that C. difficile RT 017 could be transmitted between humans and animals, and also reported that deletions and insertions found in RT 017 genomes were distributed throughout all geographical areas [Citation33]. The finding of little genetic diversity implies that C. difficile RT 017 originated in a single geographical area and that global spread occurred relatively recently, however, it remained unclear where that single geographical area was. Cairns et al. [Citation33] concluded that C. difficile RT 017 originated in North America and then spread to Europe, Asia and other parts of the world [Citation33]. This conclusion contradicts many epidemiological studies (see below) that, taken collectively, suggest that the origin of C. difficile RT 017 is in Asia. The Cairns et al. study included only a limited number of historic C. difficile RT 017 isolates from Asia (2 strains from Korea and 1 strain from Japan, all isolated in 1995) and a greater number of C. difficile RT 017 strains from North America (9 strains from the United States isolated from 1990 to 1996).
Global dissemination of C. difficile RT 017
Despite producing only one toxin, C. difficile RT 017 has successfully spread throughout the world with evidence of human infection in North America [Citation17,Citation39,Citation47,Citation63–66], Europe [Citation8,Citation13,Citation16,Citation20,Citation22,Citation23,Citation67,Citation68], Asia [Citation9,Citation14,Citation15,Citation19,Citation69–76], South America [Citation18], Africa [Citation77], and Australia [Citation78–81]. summarizes chronologically the major events surrounding the detection of C. difficile RT 017 from around the world, comparing studies of prevalence during outbreaks to studies in non-outbreak settings.
Figure 5. Timeline of C. difficile RT 017 reports around the world. Outbreaks refer to an increase in the regional prevalence of RT 017, which is confirmed either to be clonal or with evidence suggesting that isolates came from the same source. Endemic presence refers to prevalence reports that were not associated with outbreaks.
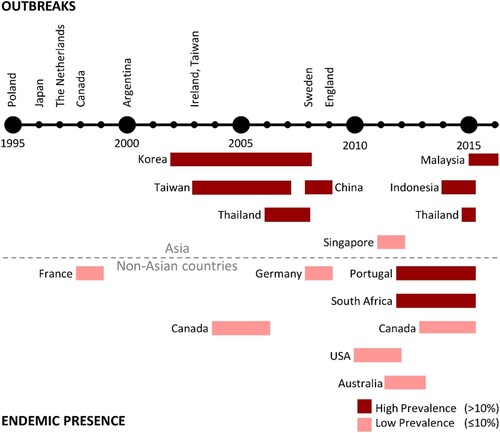
Reports on C. difficile RT 017 infection started in the late 1990s with a series of outbreaks in Poland [Citation13], Japan [Citation14,Citation15], the Netherlands [Citation16], Canada [Citation17], and Argentina [Citation18]. During the early 2000s, there were outbreaks of so-called “hypervirulent” C. difficile RT 027 in Europe and North America [Citation82], and the number of RT 017 reports appeared to subside [Citation39,Citation63,Citation67,Citation68,Citation83–85]. Still, there were further outbreaks of RT 017 infection in Ireland and Taiwan during 2003 and 2004 [Citation19–21], and in Sweden in 2008 [Citation23]. The most recent documented outbreak of RT 017 infection started in 2009 in England and persisted for at least 3 years [Citation22].
Among these C. difficile RT 017 outbreaks, clinical characteristics of the patients were described only in reports from the outbreak in Canada, with 18.8% (3/16) of cases having PMC, 31.3% (5/16) of cases being recurrent and a 37.5% (6/16) mortality rate [Citation17,Citation47]. Outbreaks of C. difficile RT 017 infection have been linked to the use of clindamycin [Citation16] and fluoroquinolones [Citation21]. In both outbreaks, discontinuation of the offending agent resulted in a rapid decline in the number of CDI cases due to C. difficile RT 017 [Citation16,Citation21]. This suggests that these outbreaks were associated with the use of specific antimicrobials and that antimicrobial stewardship helped to control spread.
Besides many outbreaks, there have also been non-outbreak reports of C. difficile RT 017 throughout the world. The majority of these reports with high prevalence figures were from Asia, while reports from non-Asian countries mostly recorded low prevalence figures. Data summarizing the prevalence of C. difficile RT 017 in Asia and non-Asian countries can be found in Tables S1 and S2 in the supplementary document.
C. difficile RT 017 in Asia
It is likely that C. difficile RT 017 is endemic in Asia and has been resident in this region for a long time, for three different reasons. First, in contrast to non-Asian countries, RT 017 appeared mainly in non-outbreak-related prevalence studies [Citation41,Citation53,Citation54,Citation69,Citation71–75,Citation86–88]. Second, there have been reports of A-B+CDT- C. difficile RTs in the region other than C. difficile RT 017 with similar deletions in the tcdA gene, some of which have also been classified in MLST clade 4 [Citation46,Citation58,Citation60,Citation61,Citation71,Citation89]. Third, the earliest Asian isolates of RT 017 in humans can be dated back to 1993 in Indonesia, where five strains of RT 017 were isolated from healthy infants [Citation15]. The high prevalence and diversity of A-B+CDT- C. difficile in Asia and the evidence of old C. difficile RT 017 isolates suggest that the origin of this RT is in Asia. While Asia is a very large continent, current information suggests that C. difficile RT 017 is endemic in at least two different regions of the continent: parts of East Asia, and South East Asia [Citation90].
East Asia
East Asia can be geographically divided into Japan and the mainland section which consists of China (including Hong Kong), North and South Korea, and the island of Taiwan. The prevalence of different C. difficile RTs in these two areas varies with RT 017 being a predominant strain only in the mainland section plus Taiwan [Citation9,Citation19,Citation69–75]. Historically, RT 017 has been responsible for ca. 15–40% of patients with CDI in South Korea [Citation9,Citation69–71], China [Citation72–74], and Taiwan [Citation19,Citation75]. In Taiwan, there was an increase in the prevalence of C. difficile RT 017 that resembled an outbreak in 2004 (73.3%; 11/15), but the prevalence eventually decreased to an endemic rate of 23.9% (11/46) in 2007 [Citation19].
In contrast to these reports, Japan saw an outbreak of C. difficile RT 017 infection in 1996 [Citation14,Citation15], perhaps coincidentally, around the same time as RT 017 outbreaks in Poland, the Netherlands and Canada [Citation13,Citation16,Citation17,Citation67]. However, there have been no major reports of C. difficile RT 017 infection in Japan since. Interestingly, in 2001, there was an outbreak of CDI caused by an A-B+ strain of C. difficile with an RT pattern resembled C. difficile RT 017 [Citation91]. This strain was later identified as the novel C. difficile RT 369, a strain that is closely related to C. difficile RT 017 [Citation36], and that was recently identified in China as ST 81, a single loci variant of ST 37 [Citation92]. To date, RT 369 remains among the most common toxigenic strains isolated in Japan while only a small number of C. difficile strains belonging to RT 017 have been detected [Citation93].
South East Asia
Most epidemiological studies in South East Asia have been conducted in Thailand [Citation41,Citation53,Citation89] with additional reports from Indonesia [Citation54], Laos [Citation86], Malaysia [Citation56,Citation87] and Singapore [Citation88]. Although the information is limited, based on these publications, and some publications from Thailand that detected a high prevalence of A-B+ C. difficile [Citation94–96], it is likely that RT 017 is endemic throughout this region.
Despite isolating C. difficile RT 017 strains as early as 1993 [Citation15], there were no epidemiological studies in the region until 2006 [Citation41]. All studies thereafter reported similar results. In Thailand, three studies confirmed that C. difficile RT 017 ranks among the most common toxigenic strains present (ca. 30.8% – 41.5%) [Citation41,Citation53,Citation89]. In Indonesia, C. difficile RT 017 was the most prevalent RT isolated from patients [Citation54]. C. difficile RT 017 has been isolated in Laos [Citation86], although only five patients were included in this report. The most recent report from South East Asia came from Malaysia where the prevalence of C. difficile RT 017 was 20.0% [Citation56]. In contrast to other South East Asian countries, a study in Singapore reported a low prevalence of RT 017 of 4.9% (3/61), and an RT distribution more like European countries. The comment was made that this possibly reflected the international population of Singapore, both resident and passing through [Citation88].
C. difficile RT 017 in non-Asian countries
Outside Asia, C. difficile RT 017 is mostly associated with outbreaks. The first group of outbreaks was reported from 1995 to 1998 in Poland [Citation13], the Netherlands [Citation16] and Canada [Citation17]. These outbreaks occurred during the same time-frame as the Japanese outbreak [Citation14,Citation15]. Since 2000, there have been four outbreaks of C. difficile RT 017 infection outside Asia [Citation18,Citation20–23]. Even though there have been non-outbreak reports of RT 017 in some parts of the world, the prevalence is low in most areas (≤10%) when compared to Asia [Citation8,Citation13,Citation16,Citation20,Citation22,Citation39,Citation63,Citation67,Citation68,Citation78–81].
North America
After 2002, C. difficile RT 017 was rapidly overshadowed by the emergence of the “hyper-virulent” C. difficile RT 027 in this region [Citation82]. The prevalence of C. difficile RT 017 in Canada decreased from 5.4% (58/1,080) during 2004–2006 [Citation63] to 1.3% (17/1,310) during 2013–2015 [Citation83]. The prevalence of C. difficile RT 017 in the United States was ca. 2–3% during 2010–2012 [Citation64–66]. In 2011, the overall prevalence of RT 017 in North America was reported at 4.3% (15/350) of toxigenic strains [Citation39].
Europe
Apart from obvious outbreaks, reports of RT 017 in Europe were scarce. During the late 1990s, the prevalence of RT 017 was 2.5% (9/364) in France [Citation67]. During 2008–2009, RT 017 was responsible for 4.9% (2/41) of severe CDI cases in Germany [Citation84]. In 2012, only one out of 171 (0.6%) C. difficile isolates from Austria was classified as C. difficile RT 017 [Citation68]. A pan-European study reported an overall prevalence of C. difficile RT 017 during 2011–2014 of 1.8% (16/866) [Citation85]. Portugal was the only European country to report a prevalence of C. difficile RT 017 higher than 10% [Citation97].
Australia
Several epidemiological studies conducted in various regions of Australia with C. difficile RT 017 being found at a much lower prevalence compared to Asia. The prevalence of C. difficile RT 017 infection was ca. 3% [Citation78–81] suggesting that those cases are are more likely to be imported rather than caused by endemic strains.
Africa
The number of studies on CDI in Africa is very limited. To date, the only country with reported C. difficile RT 017 infection is South Africa, where a very high prevalence of RT 017 among diarrhoeal patients in tuberculosis hospitals was seen [Citation77,Citation98,Citation99]. Historically, Cape Town in South Africa has been an important port city where ships coming from and going to Asia, Australia and Europe stopped during their voyages. The introduction of C. difficile RT 017 may merely reflect travel between these regions, however, it appears that C. difficile RT 017 has now become established within the hospital system in South Africa. Patients testing positive for C. difficile are at high risk of mortality, and tuberculosis is an additional risk factor for CDI in populations with HIV [Citation100].
C. difficile RT 017 in animals
Recently, many C. difficile strains associated with CDI in humans have also been isolated from animals or animal products suggesting that CDI may be transmitted from animals [Citation101]. Despite its high prevalence in the Asian human population [Citation102], there have never been any reports of C. difficile RT 017 in animals in this region [Citation103,Citation104], and it has rarely been reported in animals elsewhere. C. difficile RT 017 has been isolated from calves in Canada [Citation105] and rabbits in Italy [Citation106]. The WGS study undertaken in the United Kingdom by Cairns et al. involving 277 C. difficile RT 017 strains only included 24 strains of animal origin [Citation33]. The reasons why RT 017 is apparently not prevalent in animals have not been elucidated.
Role of antimicrobial resistance in the outbreaks of C. difficile RT 017
AMR plays an important role in the dissemination of many C. difficile RTs. Being resistant to antimicrobials while the intestinal microbiota is disrupted allows C. difficile to survive, produce toxins and eventually cause disease [Citation2]. Furthermore, being intrinsically resistant to alcohol and desiccation, C. difficile as a spore can survive within the hospital environment and spread to patients. Antimicrobial resistance has been associated with CDI outbreaks in the past; in particular, the outbreaks of “epidemic” C. difficile RT 027 in North America and Europe were associated with fluoroquinolone and rifampicin resistance.
Outbreaks of infection with C. difficile RT 017 have been linked with clindamycin- and fluoroquinolone-resistant strains [Citation13,Citation16,Citation18,Citation21]. Besides these antimicrobials, C. difficile RT 017 also has higher rates of resistance to tetracyclines and rifaximin [Citation107–109]. Tetracycline resistance was associated with an outbreak of C. difficile RT 078 [Citation110,Citation111]. Rifaximin is a derivative of rifampicin which was also associated with the outbreak of C. difficile RT 027 [Citation112,Citation113]. There is no doubt that misuse of these antimicrobials may lead to the future outbreaks of C. difficile RT 017, given that it is endemic in East and South East Asia, where tetracycline and rifampicin are commonly prescribed for many tropical infections and tuberculosis, respectively.
Conclusions
C. difficile RT 017 is one of the most successful RTs of C. difficile in the world. It was the first A-B+ C. difficile shown to cause CDI following several outbreaks. This discovery led to a better understanding of the pathogenesis of CDI in general, together with the roles of TcdA and TcdB, and eventually lead to changes in the way the laboratory diagnosis of CDI was made. The high rate of resistance to many antimicrobial agents provides hints as to how C. difficile RT 017 spread throughout the globe. It also gives us a warning that antimicrobial stewardship is needed to prevent further outbreaks.
The ancestral home of C. difficile RT 017 remains controversial, however, the weight of epidemiological evidence suggests that this strain originated in Asia and spread to other regions of the world long before the much-publicised spread of RT 027. Particular clinical characteristics of C. difficile RT 017 infection have yet to be determined. Why C. difficile RT 017 is not found more commonly in animals despite successful human spread also remains unclear, however, this may just reflect a lack of animal studies in Asia. Also, there has been no study comparing phenotypic characteristics of C. difficile RT 017, such as sporulation, germination and motility, with other epidemic strains. Since these properties are related to the spread of C. difficile, such studies may uncover important factors that help in the control of C. difficile RT 017 spread and prevent further outbreaks.
Supplemental Material
Download MS Word (71.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Squire MM, Riley TV. Clostridium difficile infection in humans and piglets: a ‘One Health’ opportunity. Curr Top Microbiol Immunol. 2013;365:299–314.
- Leffler DA, Lamont JT. Clostridium difficile Infection. N Engl J Med. 2015;373:287–288.
- Chumbler NM, Rutherford SA, Zhang Z, et al. Crystal structure of Clostridium difficile toxin A. Nat Microbiol. 2016;1:15002. doi: 10.1038/nmicrobiol.2015.2
- Rupnik M. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease? Clin Microbiol Infect. 2007;13:457–459. doi: 10.1111/j.1469-0691.2007.01687.x
- Bauer MP, Notermans DW, van Benthem BH, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4
- Stubbs SL, Brazier JS, O'Neill GL, et al. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37:461–463.
- Fawley WN, Knetsch CW, MacCannell DR, et al. Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One. 2015;10(2):e0118150. doi: 10.1371/journal.pone.0118150
- van den Berg RJ, Claas EC, Oyib DH, et al. Characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates from outbreaks in different countries by amplified fragment length polymorphism and PCR ribotyping. J Clin Microbiol. 2004;42:1035–1041. doi: 10.1128/JCM.42.3.1035-1041.2004
- Kim J, Kim Y, Pai H. Clinical characteristics and treatment outcomes of Clostridium difficile infections by PCR ribotype 017 and 018 strains. PLoS One. 2016;11:e0168849. doi: 10.1371/journal.pone.0168849
- Goorhuis A, Debast SB, Dutilh JC, et al. Type-specific risk factors and outcome in an outbreak with 2 different Clostridium difficile types simultaneously in 1 hospital. Clin Infect Dis. 2011;53:860–869. doi: 10.1093/cid/cir549
- Walker AS, Eyre DW, Wyllie DH, et al. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis. 2013;56:1589–1600. doi: 10.1093/cid/cit127
- Kim J, Pai H, Seo MR, et al. Clinical and microbiologic characteristics of tcdA-negative variant Clostridium difficile infections. BMC Infect Dis. 2012;12:109. doi: 10.1186/1471-2334-12-109
- Pituch H, van den Braak N, van Leeuwen W, et al. Clonal dissemination of a toxin-A-negative/toxin-B-positive Clostridium difficile strain from patients with antibiotic-associated diarrhea in Poland. Clin Microbiol Infect. 2001;7:442–446. doi: 10.1046/j.1198-743x.2001.00312.x
- Kato H, Kato N, Watanabe K, et al. Analysis of Clostridium difficile isolates from nosocomial outbreaks at three hospitals in diverse areas of Japan. J Clin Microbiol. 2001;39:1391–1395. doi: 10.1128/JCM.39.4.1391-1395.2001
- Rupnik M, Kato N, Grabnar M, et al. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J Clin Microbiol. 2003;41:1118–1125. doi: 10.1128/JCM.41.3.1118-1125.2003
- Kuijper EJ, de Weerdt J, Kato H, et al. Nosocomial outbreak of Clostridium difficile-associated diarrhoea due to a clindamycin-resistant enterotoxin A-negative strain. Eur J Clin Microbiol Infect Dis. 2001;20:528–534. doi: 10.1007/s100960100550
- al-Barrak A, Embil J, Dyck B, et al. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can Commun Dis Rep. 1999;25:65–69.
- Goorhuis A, Legaria MC, van den Berg RJ, et al. Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires, Argentina. Clin Microbiol Infect. 2009;15(12):1080–1086. doi: 10.1111/j.1469-0691.2009.02759.x
- Chia JH, Lai HC, Su LH, et al. Molecular epidemiology of Clostridium difficile at a medical center in Taiwan: persistence of genetically clustering of A(-)B(+) isolates and increase of A(+)B(+) isolates. PLoS One. 2013;8(10):e75471. doi: 10.1371/journal.pone.0075471
- Drudy D, Harnedy N, Fanning S, et al. Isolation and characterisation of toxin A-negative, toxin B-positive Clostridium difficile in Dublin, Ireland. Clin Microbiol Infect. 2007;13(3):298–304. doi: 10.1111/j.1469-0691.2006.01634.x
- Drudy D, Harnedy N, Fanning S, et al. Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile. Infect Control Hosp Epidemiol. 2007;28(8):932–940. doi: 10.1086/519181
- Cairns MD, Preston MD, Lawley TD, et al. Genomic epidemiology of a protracted hospital outbreak caused by a toxin A-negative Clostridium difficile sublineage PCR ribotype 017 strain in London, England. J Clin Microbiol. 2015;53(10):3141–3147. doi: 10.1128/JCM.00648-15
- Akerlund T, Alefjord I, Dohnhammar U, et al. Geographical clustering of cases of infection with moxifloxacin-resistant Clostridium difficile PCR-ribotypes 012, 017 and 046 in Sweden, 2008 and 2009. Euro Surveill. 2011;16(10). doi: 10.2807/ese.16.10.19813-en
- Hall IC, O'Toole E. Intestinal flora in new-born infants - with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child. 1935;49(2):390–402. doi: 10.1001/archpedi.1935.01970020105010
- Bartlett JG, Chang TW, Onderdonk AB. Will the real Clostridium species responsible for antibiotic-associated colitis please step forward? Lancet. 1978;311(8059):338. doi: 10.1016/S0140-6736(78)90118-6
- Teasley DG, Gerding DN, Olson MM, et al. Prospective randomised trial of metronidazole versus vancomycin for Clostridium-difficile-associated diarrhoea and colitis. Lancet. 1983;322(8358):1043–1046. doi: 10.1016/S0140-6736(83)91036-X
- Lyerly DM, Saum KE, MacDonald DK, et al. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47(2):349–352.
- Kato H, Kato N, Watanabe K, et al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36(8):2178–2182.
- Lyerly DM, Sullivan NM, Wilkins TD. Enzyme-linked immunosorbent assay for Clostridium difficile toxin A. J Clin Microbiol. 1983;17(1):72–78.
- Laughon BE, Viscidi RP, Gdovin SL, et al. Enzyme immunoassays for detection of Clostridium difficile toxins A and B in fecal specimens. J Infect Dis. 1984;149(5):781–788. doi: 10.1093/infdis/149.5.781
- Riegler M, Sedivy R, Pothoulakis C, et al. Clostridium difficile toxin B is more potent than toxin A in damaging human colonic epithelium in vitro. J Clin Invest. 1995;95(5):2004–2011. doi: 10.1172/JCI117885
- Johnson S, Kent SA, O'Leary KJ, et al. Fatal pseudomembranous colitis associated with a variant Clostridium difficile strain not detected by toxin A immunoassay. Ann Intern Med. 2001;135(6):434–438. doi: 10.7326/0003-4819-135-6-200109180-00012
- Cairns MD, Preston MD, Hall CL, et al. Comparative genome analysis and global phylogeny of the toxin variant Clostridium difficile PCR ribotype 017 reveals the evolution of two independent sublineages. J Clin Microbiol. 2017;55(3):865–876. doi: 10.1128/JCM.01296-16
- Elliott B, Androga GO, Knight DR, et al. Clostridium difficile infection: evolution, phylogeny and molecular epidemiology. Infect Genet Evol. 2017;49:1–11. doi: 10.1016/j.meegid.2016.12.018
- Depitre C, Delmee M, Avesani V, et al. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J Med Microbiol. 1993;38(6):434–441. doi: 10.1099/00222615-38-6-434
- Senoh M, Kato H, Fukuda T, et al. Predominance of PCR-ribotypes, 018 (smz) and 369 (trf) of Clostridium difficile in Japan: a potential relationship with other global circulating strains? J Med Microbiol. 2015;64(10):1226–1236. doi: 10.1099/jmm.0.000149
- Johnson S, Sambol SP, Brazier JS, et al. International typing study of toxin A-negative, toxin B-positive Clostridium difficile variants. J Clin Microbiol. 2003;41(4):1543–1547. doi: 10.1128/JCM.41.4.1543-1547.2003
- Rupnik M, Brazier JS, Duerden BI, et al. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology. 2001;147(Pt 2):439–447. doi: 10.1099/00221287-147-2-439
- Tenover FC, Akerlund T, Gerding DN, et al. Comparison of strain typing results for Clostridium difficile isolates from North America. J Clin Microbiol. 2011;49(5):1831–1837. doi: 10.1128/JCM.02446-10
- Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48(3):770–778. doi: 10.1128/JCM.01796-09
- Ngamskulrungroj P, Sanmee S, Putsathit P, et al. Molecular epidemiology of Clostridium difficile infection in a large teaching hospital in Thailand. PLoS One. 2015;10(5):e0127026. doi:10.1371/journal.pone.0127026.
- Cohen SH, Tang YJ, Silva J, Jr. Molecular typing methods for the epidemiological identification of Clostridium difficile strains. Expert Rev Mol Diagn. 2001;1(1):61–70. doi: 10.1586/14737159.1.1.61
- Delmee M, Homel M, Wauters G. Serogrouping of Clostridium difficile strains by slide agglutination. J Clin Microbiol. 1985;21(3):323–327.
- Kimura B. Will the emergence of core genome MLST end the role of in silico MLST? Food Microbiol. 2018;75:28–36. doi: 10.1016/j.fm.2017.09.003
- Li R, Xiao D, Yang J, et al. Identification and characterization of Clostridium difficile sequence type 37 genotype by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2018;56(5). doi: 10.1128/JCM.01990-17
- Cheng J-W, Liu C, Kudinha T, et al. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify MLST clade 4 Clostridium difficile isolates. Diagn Microbiol Infect Dis. 2018;92(1):19–24. doi: 10.1016/j.diagmicrobio.2018.04.011
- Alfa MJ, Kabani A, Lyerly D, et al. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J Clin Microbiol. 2000;38(7):2706–2714.
- Chaves-Olarte E, Low P, Freer E, et al. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J Biol Chem. 1999;274(16):11046–11052. doi: 10.1074/jbc.274.16.11046
- von Eichel-Streiber C, Zec-Pirnat I, Grabnar M, et al. A nonsense mutation abrogates production of a functional enterotoxin A in Clostridium difficile toxinotype VIII strains of serogroups F and X. FEMS Microbiol Lett. 1999;178(1):163–168. doi: 10.1016/S0378-1097(99)00327-4
- Drudy D, Fanning S, Kyne L. Toxin A-negative, toxin B-positive Clostridium difficile. Int J Infect Dis. 2007;11(1):5–10. doi: 10.1016/j.ijid.2006.04.003
- Wang R, Suo L, Chen HX, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from the Chinese People’s Liberation Army General Hospital in China. Int J Infect Dis. 2018;67:86–91. doi: 10.1016/j.ijid.2017.07.010
- Persson S, Torpdahl M, Olsen KE. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008;14(11):1057–1064. doi: 10.1111/j.1469-0691.2008.02092.x
- Putsathit P, Maneerattanaporn M, Piewngam P, et al. Prevalence and molecular epidemiology of Clostridium difficile infection in Thailand. New Microbes New Infect. 2017;15:27–32. doi: 10.1016/j.nmni.2016.10.004
- Collins DA, Gasem MH, Habibie TH, et al. Prevalence and molecular epidemiology of Clostridium difficile infection in Indonesia. New Microbes New Infect. 2017;18:34–37. doi: 10.1016/j.nmni.2017.04.006
- Thipmontree W, Kiratisin P, Manatsathit S, et al. Epidemiology of suspected Clostridium difficile-associated hospital-acquired diarrhea in hospitalized patients at Siriraj Hospital. J Med Assoc Thai. 2011;94(Suppl 1):S207–S216.
- Riley TV, Collins DA, Karunakaran R, et al. High prevalence of toxigenic and non-toxigenic Clostridium difficile in Malaysia. J Clin Microbiol. 2018;56(6):e00170–e001718. doi: 10.1128/JCM.00170-18
- Songer JG, Jones R, Anderson MA, et al. Prevention of porcine Clostridium difficile-associated disease by competitive exclusion with nontoxigenic organisms. Vet Microbiol. 2007;124(3-4):358–361. doi: 10.1016/j.vetmic.2007.04.019
- Chen YB, Gu SL, Wei ZQ, et al. Molecular epidemiology of Clostridium difficile in a tertiary hospital of China. J Med Microbiol. 2014;63(Pt 4):562–569. doi: 10.1099/jmm.0.068668-0
- Dingle KE, Elliott B, Robinson E, et al. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol. 2014;6(1):36–52. doi: 10.1093/gbe/evt204
- Qin J, Dai Y, Ma X, et al. Nosocomial transmission of Clostridium difficile genotype ST81 in a general teaching hospital in China traced by whole genome sequencing. Sci Rep. 2017;7(1):9627. doi: 10.1038/s41598-017-09878-8
- Wang X, Cai L, Yu R, et al. ICU-Onset Clostridium difficile infection in a university hospital in China: a prospective cohort study. PLoS One. 2014;9(11):e111735. doi: 10.1371/journal.pone.0111735
- He M, Sebaihia M, Lawley TD, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A. 2010;107(16):7527–7532 doi: 10.1073/pnas.0914322107
- Martin H, Willey B, Low DE, et al. Characterization of Clostridium difficile strains isolated from patients in Ontario, Canada, from 2004 to 2006. J Clin Microbiol. 2008;46(9):2999–3004. doi: 10.1128/JCM.02437-07
- Dubberke ER, Reske KA, Seiler S, et al. Risk factors for acquisition and loss of Clostridium difficile colonization in hospitalized patients. Antimicrob Agents Chemother. 2015;59(8):4533–4543. doi: 10.1128/AAC.00642-15
- Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(24):2369–2370.
- Waslawski S, Lo ES, Ewing SA, et al. Clostridium difficile ribotype diversity at six health care institutions in the United States. J Clin Microbiol. 2013;51(6):1938–1941. doi: 10.1128/JCM.00056-13
- Barbut F, Lalande V, Burghoffer B, et al. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J Clin Microbiol. 2002;40(6):2079–2083. doi: 10.1128/JCM.40.6.2079-2083.2002
- Indra A, Schmid D, Huhulescu S, et al. Clostridium difficile ribotypes in Austria: a multicenter, hospital-based survey. Wien Klin Wochenschr. 2015;127(15-16):587–593. doi: 10.1007/s00508-015-0808-5
- Kim H, Jeong SH, Roh KH, et al. Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J Lab Med. 2010;30(5):491–497. doi: 10.3343/kjlm.2010.30.5.491
- Kim J, Kang JO, Kim H, et al. Epidemiology of Clostridium difficile infections in a tertiary-care hospital in Korea. Clin Microbiol Infect. 2013;19(6):521–527. doi: 10.1111/j.1469-0691.2012.03910.x
- Kim H, Riley TV, Kim M, et al. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J Clin Microbiol. 2008;46(3):1116–1117. doi: 10.1128/JCM.01188-07
- Huang H, Fang H, Weintraub A, et al. Distinct ribotypes and rates of antimicrobial drug resistance in Clostridium difficile from Shanghai and Stockholm. Clin Microbiol Infect. 2009;15(12):1170–1173. doi: 10.1111/j.1469-0691.2009.02992.x
- Huang H, Weintraub A, Fang H, et al. Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe. 2010;16(6):633–635. doi: 10.1016/j.anaerobe.2010.09.002
- Jin D, Luo Y, Huang C, et al. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in Eastern China. J Clin Microbiol. 2017;55(3):801–810. doi: 10.1128/JCM.01898-16
- Hung YP, Huang IH, Lin HJ, et al. Predominance of Clostridium difficile ribotypes 017 and 078 among toxigenic clinical isolates in Southern Taiwan. PLoS One. 2016;11(11):e0166159. doi: 10.1371/journal.pone.0166159
- Chen YB, Gu SL, Shen P, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from hospitals during a 4-year period in China. J Med Microbiol. 2018;67(1):52–59. doi: 10.1099/jmm.0.000646
- Kullin B, Brock T, Rajabally N, et al. Characterisation of Clostridium difficile strains isolated from Groote Schuur Hospital, Cape Town, South Africa. Eur J Clin Microbiol Infect Dis. 2016;35(10):1709–1718. doi: 10.1007/s10096-016-2717-6
- Collins DA, Putsathit P, Elliott B, et al. Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012. Pathology. 2017;49(3):309–313. doi: 10.1016/j.pathol.2016.10.013
- Eyre DW, Tracey L, Elliott B, et al. Emergence and spread of predominantly community-onset Clostridium difficile PCR ribotype 244 infection in Australia, 2010 to 2012. Euro Surveill. 2015;20(10):21059. doi: 10.2807/1560-7917.ES2015.20.10.21059
- Knight DR, Giglio S, Huntington PG, et al. Surveillance for antimicrobial resistance in Australian isolates of Clostridium difficile, 2013–14. J Antimicrob Chemother. 2015;70(11):2992–2999. doi: 10.1093/jac/dkv220
- Foster NF, Collins DA, Ditchburn SL, et al. Epidemiology of Clostridium difficile infection in two tertiary-care hospitals in Perth, Western Australia: a cross-sectional study. New Microbes New Infect. 2014;2(3):64–71. doi: 10.1002/nmi2.43
- Warny M, Pepin J, Fang A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366(9491):1079–1084. doi: 10.1016/S0140-6736(05)67420-X
- Karlowsky JA, Adam HJ, Kosowan T, et al. PCR ribotyping and antimicrobial susceptibility testing of isolates of Clostridium difficile cultured from toxin-positive diarrheal stools of patients receiving medical care in Canadian hospitals: the Canadian Clostridium difficile Surveillance Study (CAN-DIFF) 2013–2015. Diagn Microbiol Infect Dis. 2018;91(2):105–111. doi: 10.1016/j.diagmicrobio.2018.01.017
- Arvand M, Hauri AM, Zaiss NH, et al. Clostridium difficile ribotypes 001, 017, and 027 are associated with lethal C. difficile infection in Hesse, Germany. Euro Surveill. 2009;14(45). doi: 10.2807/ese.14.45.19403-en
- Freeman J, Vernon J, Morris K, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21(3):248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017
- Cheong E, Roberts T, Rattanavong S, et al. Clostridium difficile infection in the Lao People’s Democratic Republic: first isolation and review of the literature. BMC Infect Dis. 2017;17(1):635. doi: 10.1186/s12879-017-2737-6
- Zainul NH, Ma ZF, Besari A, et al. Prevalence of Clostridium difficile infection and colonization in a tertiary hospital and elderly community of North-Eastern Peninsular Malaysia. Epidemiol Infect. 2017;145(14):3012–3019. doi: 10.1017/S0950268817002011
- Tan XQ, Verrall AJ, Jureen R, et al. The emergence of community-onset Clostridium difficile infection in a tertiary hospital in Singapore: a cause for concern. Int J Antimicrob Agents. 2014;43(1):47–51. doi: 10.1016/j.ijantimicag.2013.09.011
- Imwattana K, Wangroongsarb P, Riley TV. High prevalence and diversity of tcdA-negative and tcdB-positive, and non-toxigenic, Clostridium difficile in Thailand. Anaerobe. 2019;57:4–10. doi: 10.1016/j.anaerobe.2019.03.008
- Collins DA, Sohn KM, Wu Y, et al., editors. Clostridium difficile infection in the Asia-Pacific region. 27th European Congress of Clinical Microbiology and Infectious Diseases; 2017; Vienna, Austria.
- Sato H, Kato H, Koiwai K, et al. A nosocomial outbreak of diarrhea caused by toxin A-negative, toxin B-positive Clostridium difficile in a cancer center hospital. Kansenshogaku Zasshi. 2004;78(4):312–319. doi: 10.11150/kansenshogakuzasshi1970.78.312
- Wang B, Peng W, Zhang P, et al. The characteristics of Clostridium difficile ST81, a new PCR ribotype of toxin A- B+ strain with high-level fluoroquinolones resistance and higher sporulation ability than ST37/PCR ribotype 017. FEMS Microbiol Lett. 2018;365(17). doi: 10.1093/femsle/fny168
- Mikamo H, Aoyama N, Sawata M, et al. The effect of bezlotoxumab for prevention of recurrent Clostridium difficile infection (CDI) in Japanese patients. J Infect Chemother. 2018;24(2):123–129. doi: 10.1016/j.jiac.2017.10.005
- Wangroongsarb P, Kamthalang T, Jittaprasatsin C, et al. Antimicrobial susceptibility and toxin production of Clostridium difficile isolated from diarrheal patients during 2012–2015. J Assoc Med Sci. 2017;50(2):187–196.
- Chankhamhaengdecha S, Hadpanus P, Aroonnual A, et al. Evaluation of multiplex PCR with enhanced spore germination for detection of Clostridium difficile from stool samples of the hospitalized patients. Biomed Res Int. 2013;2013:875437.
- Chotiprasitsakul D, Janvilisri T, Kiertiburanakul S, et al. A superior test for diagnosis of Clostridium difficile-associated diarrhea in resource-limited settings. Jpn J Infect Dis. 2012;65(4):326–329. doi: 10.7883/yoken.65.326
- Isidro J, Santos A, Nunes A, et al. Imipenem resistance in Clostridium difficile ribotype 017, Portugal. Emerg Infect Dis. 2018;24(4):741–745. doi: 10.3201/eid2404.170095
- Kullin B, Wojno J, Abratt V, et al. Toxin A-negative toxin B-positive ribotype 017 Clostridium difficile is the dominant strain type in patients with diarrhoea attending tuberculosis hospitals in Cape Town, South Africa. Eur J Clin Microbiol Infect Dis. 2017;36(1):163–175. doi: 10.1007/s10096-016-2790-x
- Rajabally N, Kullin B, Ebrahim K, et al. A comparison of Clostridium difficile diagnostic methods for identification of local strains in a South African centre. J Med Microbiol. 2016;65(4):320–327. doi: 10.1099/jmm.0.000231
- Legenza L, Barnett S, Rose W, et al. Epidemiology and outcomes of Clostridium difficile infection among hospitalised patients: results of a multicentre retrospective study in South Africa. BMJ Glob Health. 2018;3(4):e000889. doi: 10.1136/bmjgh-2018-000889
- Bauer MP, Kuijper EJ. Potential sources of Clostridium difficile in human infection. Infect Dis Clin North Am. 2015;29(1):29–35. doi: 10.1016/j.idc.2014.11.010
- Collins DA, Hawkey PM, Riley TV. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control. 2013;2(1):21. doi: 10.1186/2047-2994-2-21
- Cho A, Byun JW, Kim JW, et al. Low prevalence of Clostridium difficile in slaughter pigs in Korea. J Food Prot. 2015;78(5):1034–1036. doi: 10.4315/0362-028X.JFP-14-493
- Wu Y-C, Chen C-M, Kuo C-J, et al. Prevalence and molecular characterization of Clostridium difficile isolates from a pig slaughterhouse, pork, and humans in Taiwan. Int J Food Microbiol. 2017;242:37–44. doi: 10.1016/j.ijfoodmicro.2016.11.010
- Rodriguez-Palacios A, Stampfli HR, Duffield T, et al. Clostridium difficile PCR ribotypes in calves, Canada. Emerg Infect Dis. 2006;12(11):1730–1736. doi: 10.3201/eid1211.051581
- Drigo I, Mazzolini E, Bacchin C, et al. Molecular characterization and antimicrobial susceptibility of Clostridium difficile isolated from rabbits raised for meat production. Vet Microbiol. 2015;181(3–4):303–307. doi: 10.1016/j.vetmic.2015.10.005
- Putsathit P, Maneerattanaporn M, Piewngam P, et al. Antimicrobial susceptibility of Clostridium difficile isolated in Thailand. Antimicrob Resist Infect Control. 2017;6:58. doi: 10.1186/s13756-017-0214-z
- Huang H, Weintraub A, Fang H, et al. Antimicrobial resistance in Clostridium difficile. Int J Antimicrob Agents. 2009;34(6):516–522. doi: 10.1016/j.ijantimicag.2009.09.012
- Chow VCY, Kwong TNY, So EWM, et al. Surveillance of antibiotic resistance among common Clostridium difficile ribotypes in Hong Kong. Sci Rep. 2017;7(1):17218. doi: 10.1038/s41598-017-17523-7
- Bakker D, Corver J, Harmanus C, et al. Relatedness of human and animal Clostridium difficile PCR ribotype 078 isolates determined on the basis of multilocus variable-number tandem-repeat analysis and tetracycline resistance. J Clin Microbiol. 2010;48(10):3744–3749. doi: 10.1128/JCM.01171-10
- Dingle KE, Didelot X, Quan TP, et al. A role for tetracycline selection in the evolution of Clostridium difficile PCR-ribotype 078. bioRxiv. 2018;262352.
- He M, Miyajima F, Roberts P, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45(1):109–113. doi: 10.1038/ng.2478
- Curry SR, Marsh JW, Shutt KA, et al. High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin Infect Dis. 2009;48(4):425–429. doi: 10.1086/596315
