ABSTRACT
Pertussis caused by Bordetella pertussis, remains a public health problem worldwide, despite high vaccine coverage in infants and children in many countries. Iran has been using whole cell vaccine for the last 50 years with more than 95% vaccination rate since 1988 and has experienced pertussis resurgence in recent years. Here, we sequenced 55 B. pertussis isolates mostly collected from three provinces with the highest number of pertussis cases in Iran, including Tehran, Mazandaran, and Eastern-Azarbayjan from the period of 2008-2016. Most isolates carried ptxP3/prn2 alleles (42/55, 76%), the same genotype as isolates circulating in acellular vaccine-administrating countries. The second most frequent genotype was ptxP3/prn9 (8/55, 14%). Only three isolates (5%) were ptxP1. Phylogenetic analysis showed that Iranian ptxP3 isolates can be divided into eight clades (Clades 1-8) with no temporal association. Most of the isolates from Tehran grouped together as one distinctive clade (Clade 8) with six unique single nucleotide polymorphisms (SNPs). In addition, the prn9 isolates were grouped together as Clade 5 with 12 clade-supporting SNPs. No pertactin deficient isolates were found among the 55 Iranian isolates. Our findings suggest that there is an ongoing adaptation and evolution of B. pertussis regardless of the types of vaccine used.
Introduction
Despite high vaccine coverage in many countries, pertussis, also known as whooping cough is still reported in all age groups with the highest severity in infants and young children [Citation1–3]. Pathogen adaptation through genetic changes has been shown to be one of the key roles in the re-emergence of pertussis, especially in countries were acellular vaccines (ACV) were used which commonly contained three (pertussis toxin (Ptx), pertactin (Prn), and filamentous haemagglutinin (Fha)) or five components (additional fimbriae Fim2 and Fim3) [Citation1,Citation3,Citation4–6]. It has been shown that currently predominant B. pertussis isolates in many countries harbour non-ACV-antigen encoding alleles. The most common allelic profile worldwide, ptxA1/ptxP1/prn1, has changed to ptxA1/ptxP3/prn2 [Citation7]. ptxP3, the pertussis toxin (Ptx) promoter allele has been shown to be associated with higher virulence [Citation5] and ptxP3 isolates have a better fitness in animal studies [Citation8].
While whole cell vaccine (WCV) has been replaced by ACV in many countries [Citation2], it is still used in Iran since the 1950s for all scheduled doses in infants at 2, 4 and 6 months of age as well as two boosters at 18 months of age and six years old children [Citation9,Citation10]. Since 1988 vaccine coverage has increased to more than 95%, pertussis cases dramatically has declined from 40 cases per 100,000 in 1979 to 1.12 per 100,000 of population in 2011 [Citation11,Citation12]. However, increased pertussis notification rate has been reported in Iran since 2007 peaking in 2013 with 1415 suspected cases per 100,000 during the 2012–2014 pertussis epidemic [Citation13–15]. The introduction of PCR based diagnosis, incomplete vaccination and the new guidelines and policies implemented in 2009 by Iran Centre for Communicable Disease Control that all suspected and confirmed cases must be notified and samples must be sent to the reference laboratory may have contributed to the increased number of reported pertussis cases in Iran [Citation16].
In this study we analysed a selected set of B. pertussis isolates from 2008 to 2016 from nine provinces in Iran, especially from provinces with high number of pertussis cases to determine the evolutionary characteristics of Iranian B. pertussis using whole genome sequencing (WGS). Our aim was to expand our knowledge of the evolutionary forces, the genetic loci undergoing selective pressure and how B. pertussis may be evolving in response to WCV vaccine pressure in a country where WCV has been used in the last 50 years.
Methods and materials
Bacterial isolates and whole genome sequencing
A total of 154 clinical isolates from 2008 to 2016 from different provinces of Iran were available in Pasteur Institute of Iran as a reference diagnostic laboratory ( (A)). Most of these isolates (107/154, 69.48%) were from patients aged up to 6-month-old and 40.26% (62/154), 32.46% (50/154) and 27.27% (42/154) were unvaccinated, partially or fully vaccinated respectively. Isolates were collected from 24 out of 31 provinces of which three provinces had more than 20 isolates including Eastern-Azarbayjan, Tehran and Mazandaran with 32, 29 and 23 isolates respectively.
Figure 1. Basic statistics of isolates studied. (A) Distribution of isolates by year and vaccination status. A total of 154 B. pertussis isolates were available from the Reference laboratory. The graph shows their distribution by year and the proportion by the age of patients during 2008–2016. (B) The distribution of selected isolates and their ACV antigen gene profile results from different provinces. P1: Eastern-Azarbayjan, P4: Esfahan, P8: Tehran, P9: Charmahal, P27: Mazandaran. Numbers in the parentheses represent the number of selected isolates out of the total number of isolates available for each province.
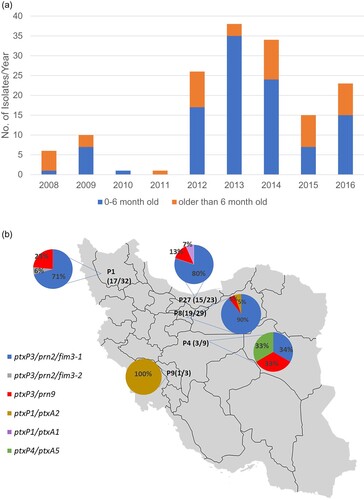
We selected a total of 55 currently circulating Iranian B. pertussis isolates from 2008 to 2016 based on year and province of isolation. Amongst these 55 B. pertussis isolates, 51 were selected from three provinces which had the highest number of collected samples, including Tehran (Province-8), Mazandaran (northern province, Province-27) and Eastern-Azarbayjan (north-eastern province, Province-1) with 19, 15 and 17 isolates, respectively ((B)). Thirty-two of the 51 isolates were from the 2012–2014 epidemic (Supplementary File-1). Additionally, four isolates from two provinces located in the centre of Iran (IR141, IR142, IR164 from Esfahan (Province-4), and IR138 from Charmahal (Province-9)) were added to have a better picture of geographical relationship of the isolates ((B)).
DNA was extracted from bacterial culture as described previously [Citation17]. The genomes were sequenced using 150 bp paired-end Illumina NextSeq sequencing with a minimum coverage of 150-fold. The raw reads were submitted to GeneBank under BioProject No. PRJNA450302.
Data analysis
The distribution of Insertion sequence (IS) elements and SNP calling was performed and variation in genes encoding ACV antigens (prn, ptxA, ptxP, fim2 and fim3) was investigated as described previously [Citation17]. We detected SNPs and short insertion and deletions (indels) with less than 100 bp as previously described using Burrows–Wheeler Alignment (BWA) tools (v0.7.12), SAMtools (v0.1.19) and progressiveMauve (v2.3.1) [Citation17–20]. Prokka (v1.12) and Roary (v3.11.2) were used for genome annotation and defining the core genomes, respectively [Citation21,Citation22]. de novo assembly was performed for all sequencing data to assemble reads into contigs using SPAdes (v3.7.0) [Citation23]. Contigs were also aligned to B. pertussis strains Tohama I (GenBank Accession No. BX470248) and CS (Accession No. CP002695) as references to generate comparable data using progressiveMauve [Citation18].
Phylogenetic analysis was conducted using MEGA (v7) [Citation24]. The maximum parsimony algorithm was used to construct phylogenetic tree. Tree-Bisection-Reconnection (TBR) was used to search optimal trees. Bootstrap analysis was based on 500 replicates and B. pertussis Tohama I used as outgroup.
Results
ACV vaccine antigen gene allele profiles
Typing of ACV antigen genes, ptxP, ptxA, prn, fim2 and fim3 among the 55 selected isolates from 2008 to 2016 revealed that ptxP3/ptxA1/fim2-1 isolates were predominant (51/55, 92.7%). Out of the 51 ptxP3 isolates, 50 isolates carried fim3-2 (also denoted as fim3B) allele and only one had fim3-1 (also denoted as fim3A) [Citation25,Citation26]. Isolates with the allele profile ptxP3/ptxA1/prn2/fim3- 1were predominant among ptxP3 isolates (42/51, 82.3%). We found a new profile of ptxP3/ptxA1/prn9/fim3-2 with eight isolates (8/51,15.68%) as the second most frequent profile. All prn9 isolates were isolated after 2013 and rose from 12.5% (2/15) in 2013 to 27% (3/11) in 2016. These isolates were not reported previously in Iran and collected from different provinces showing the expansion of prn9 isolates within the country ((B)). There were only four non-ptxP3 isolates, one, IR142, harboured a ptxP4 allele (full profile: ptxP4/ptxA5/prn6/ fhaB2/ fim2-1/ fim3-1), and three were ptxP1 which were further differentiated by ptxA allele into ptxP1/ptxA1/fim2-1/fim3-1/ prn1 (one isolates, IR146) and ptxP1/ptxA2/fim2-1/ fim3-1/ prn1 (two isolates, IR136 and IR138).
Phylogeny of Iranian B. pertussis isolates
Phylogenetic analysis based on SNPs across the whole-genome differentiated the 55 isolates into three major lineages, 51 ptxP3 isolates into one lineage, three ptxP1 isolates into the second () and the outlier (IR142 with ptxP4/ ptxA5/ prn6/ fhaB2 allele profile) which were omitted from the tree due to high number of SNPs and will be discussed later. There were 32 SNPs shared by all ptxP3 Iranian isolates () but none was unique to Iranian isolates, some of which were also reported among global non-ptxP3 isolates [Citation7]. All except one ptxP3 isolate were grouped together as one lineage and all carried fim3-2 allele with 7 SNPs supporting the node and were further divided into eight clades as Clade 1 to Clade 8 supported by 1, 10, 6, 19, 12, 8, 5 and 6 SNPs, respectively ( and ). There was no correlation between isolates in terms of year of isolation except for Clade 3 and Clade 4. There were only 2 isolates each in Clade 3 and Clade 4 which were obtained in 2013 and 2014, respectively. The 2012–2014 epidemic isolates were interspersed within and between clades with no temporal correlation. Interestingly, all eight prn9 isolates were clustered as Clade 5 supported by 12 unique SNPs. Clade 8, supported by 6 SNPs, was the largest clade with 20 isolates, of which 12 (70%) were from Tehran. One isolate (IR87) collected in 2013 from Eastern-Azarbayjan was not grouped within any clades and defined as an ungrouped isolate. The two ptxP1/ptxA2 isolates were collected in 2015 and were well separated from the ptxP1/ptxA1 isolate ().
Figure 2. Maximum parsimony tree of the 54 Iranian B. pertussis isolates. The tree was constructed based on 653 SNPs. The number on the internal and terminal branches corresponds to the number of SNPs supporting each branch. Iranian ptxP3/ fim3-2 isolates were grouped into 8 clades as marked. The majority of ptxP3 isolates collected from Tehran grouped together (12 out of 18) in clade 8. New profile of ptxP3/ ptxA1/prn9 was defined as clade 5. Black circle on the tree represents the distribution of isolates collected from Tehran.
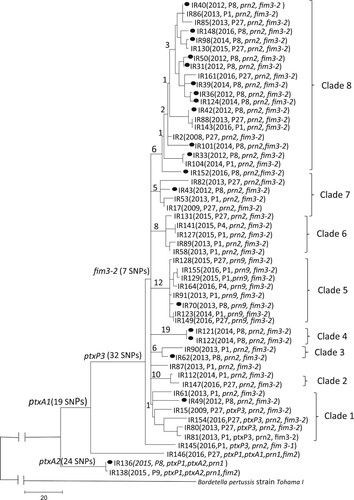
Table 1. Single nucleotide polymorphism (SNP) and short insertion and deletions (indels) supporting the nodes and clades in phylogenetic analysis.
SNPs or indels associated with virulence genes or potential adaptive evolution
We examined the SNPs and indels that may be associated with gene functions and play an adaptive role. From 654 SNPs found in this study (Supplementary File-2), a total of 310 SNPs were unique to one or more Iranian isolates which had not been reported previously. Nine nsSNP were from virulence associated genes including sphB2, sphB3, tcfA, bscQ, vag8, fhaL, ompQ, brkA and ptxB genes (). There are 11 nonsense (stop codon) SNPs, eight of which were unique to Iranian isolates (). A novel nonsense SNP in fhaD was found in IR136 and IR138, both of which were ptxP1/ptxA2 isolates. fhaD encoding a chaperone protein is known to be an accessory gene which may affect the production and secretion of Fha and fimbriae in B. pertussis [Citation27].
Table 2. Unique non-synonymous SNPs in virulence associated genes or nonsense SNPs that were only found in Iranian B. pertussis isolates.
Some clade dividing SNPs may be clade specific adaptive changes including changes in the promoter or coding region of virulence associated genes or metabolic genes. Three of the 12 SNPs supporting the grouping of prn9 isolates as Clade 5 () were of interest: one SNP located in the promoter region of the dnt (BP3439) gene encoding dermonecrotic toxin, one in the promoter region of fhaB and bvgA genes and the third a nsSNP located in BP2548 encoding a histidine kinase. The mutation in the fhaB promoter region was also found in the Fha/Prn negative B. pertussis strain isolated in 2009 in France [Citation28]. However, it remains unknown if this mutation leads to non-FhaB/Prn expression since it was identified in one UK isolate (UK98) which appeared to be Fha and Prn positive [Citation7,Citation29–31]. The nsSNP in BP2548 which encodes a two-component system histidine kinase and has not been reported previously. One of the six SNPs supporting Clade 8 was a nsSNP located in the glpK gene which encodes a glycerol kinase; a key enzyme in the regulation of glycerol uptake and metabolism.
Insertion sequence elements
A total of 19 IS elements due to the IS481F or IS481R (F and R denote insertion in the forward or reverse direction) that were absent in Tohama I were identified (Supplementary File-3), all of which have been reported previously [Citation17,Citation30]. One IS in BP1446 encoding enoyl-CoA hydratase was found in all Clade 2 isolates. Two were found in Clade 4 with an IS481F in BP2232 encoding hypothetical protein and an IS481R in BP3588 encoding acetylornithine deacetylase. Furthermore, two IS481R insertions located in BP1591 and BP3764 were found only in ptxA2 isolates, IR136 and IR138. IR142 was compared with Tohama I separately and 22 IS disruptions were found for this isolate, of which, 19 were unique. It is of interest that IR142 shared with B. pertussis 18323 an IS481R in the fhaL gene encoding a putative protein homologous to Fha that led to a pseudogene [Citation32]. No IS disruptions were identified in prn or other ACV-antigen genes.
Region of differences in Iranian B. pertussis genomes
Current circulating ptxP3 isolates appear to have lost clusters of genes, also known as regions of difference (RD) including RD3 (BP0910A-BP0937), RD5 (BP1134-BP1141) and RD10 (BP1947-1968) [Citation33–35]. We found 59 loci with more than two genes deleted in one or more isolates (Supplementary File-4) including RD3 deleted in all 55 isolates while RD5 was deleted in all isolates except IR142. Only BP1134 and BP1142 of RD5 were deleted in IR142. RD10 was absent in all ptxP3 isolates whereas only BP1947 and BP1968 of RD10 were deleted in all non-ptxP3 isolates. A set of 20 genes (BP1157-BP1177), of which six encoding cell surface proteins, were deleted in all prn9 isolates and IR146 (ptxP1/ptxA2/prn1). Two large fragments, containing 18 (BPTD2835-BPTD2852) and 21 (BPTD0387 to BPTD0407) genes, that were absent in Tohama I but present in the other reference Chinese CS strain and many other isolates [Citation36,Citation37], were also present in all Iranian isolates.
The evolutionary relationships of Iranian B. pertussis from the global perspective
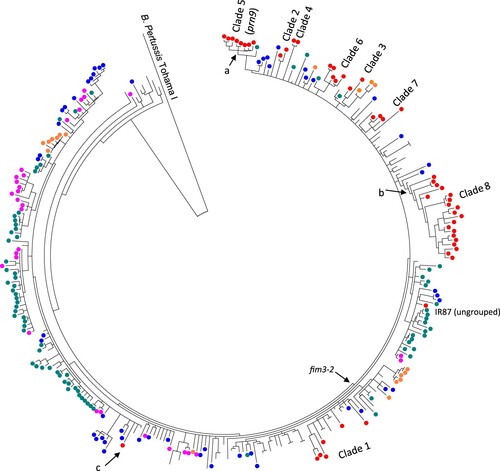
To place the Iranian isolates in a global context, we determined the genetic relationships of the 51 Iranian B. pertussis ptxP3 isolates with 252 ptxP3 genomes representing 18 countries from 1988 to 2016 from multiple studies [Citation7,Citation17,Citation29,Citation30] (). ptxP3 isolates were distributed among two large lineages demarcated by fim3 alleles as reported previously [Citation7,Citation17,Citation29]. The only fim3-1 isolate, IR145, grouped with fim3-1 isolates from the USA. Clade 1 isolates were distributed among global isolates isolated between 1997–2003 suggesting multiple parallel spread of B. pertussis isolates into Iran. Clade 2 to 8 were still clustered together with each clade being separately close to isolates mainly from the USA. prn9 isolates as Clade 5 were grouped with one prn2 isolate from the UK (UK98 isolated in 2012). From 12 unique SNPs for prn9 isolates as clade 5, seven were shared with UK98 and two nsSNPs positioned in BP2548 and BP3694 and one mutation in promoter of BP0359 were still unique to Iranian isolates. Clade 8 as largest Iranian clade grouped with one Canadian isolate (B062, isolated in 2001) and from 6 unique SNPs for this clade 1 nsSNP located in B1245 was still unique to Iranian isolates. Finally, the ungrouped isolate, IR87, was grouped with UK fim3-2 isolates.
Figure 3. Phylogenetic relationships of Iranian ptxP3 B. pertussis isolates with global isolates. The tree was inferred using maximum parsimony method using B. pertussis Tohama I as an outgroup. A total of 304 ptxP3 isolates from different continents were included (Red: Iran, Blue: The USA, Green: UK, Pink: Australia) (details of the isolates are available in Supplementary File-1). Iranian isolates showed the same clustering as clade 2–8 except isolates in clade 1 that were interspersed with isolates from other countries. Clade 5 had 12 clade supporting SNPs, seven of which were shared with UK98 prn2 isolate (green circle) and two nsSNPs positioned in BP2548 and BP3694 and one mutation in the promoter of BP0359 were still unique to Iranian isolates (Node marked with a). For 6 clade 8 specific SNPs, five were shared with one Canadian isolate (B062, isolated in 2001) and one nsSNPs located in B1245 was still unique to Iranian isolates (Node marked with b). The only ptxP3/fim3-1 Iranian isolate (IR145) was grouped with fim3-1 isolates from USA (Node marked with c).
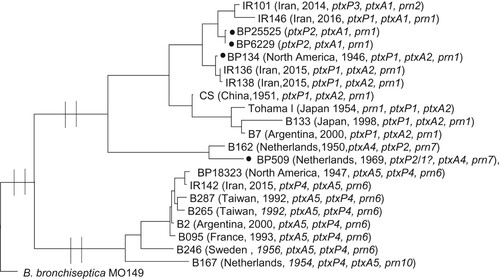
The relationship of IR142, which stands as a distinct lineage from the remaining 54 Iranian B. pertussis isolates was investigated. The phylogenetic analysis revealed that IR142 was more closely related to some old divergent isolates (seven isolates from different continents) from before 1950s reported by Bart et al. [Citation7] and in particular to B. pertussis 18323 () with the same allelic profile. Furthermore, the relationship of four strains used as WCV vaccine seeds, BP134, BP509, BP6229 and BP25525, with recently collected isolates from Iran were also investigated [Citation38]. The four strains have been used to manufacture the Indian DTwP, which has been used in Iran for immunization in recent years. BP509 and BP134 were also used as vaccine seeds from 1950 by Iran to manufacture its own DTwP [Citation39]. Four recent Iranian isolates were selected for comparison including three ptxP1 isolates (IR136, IR138 and IR146) which have almost the same allelic profile with three vaccine seed strains and one random ptxP3 isolate (IR101) which was more distinct. The results revealed that BP509 with ptxA4/prn7 genotype was more distinct than other vaccine seeds. The other three vaccine seed strains grouped together with the ptxP1 isolates including IR136, IR138 and IR146 (). As expected, all vaccine seed strains were distinct from the current circulating ptxP3 isolate (IR101).
Figure 4. Maximum Parsimony Phylogeny using 2792 SNPs to analyse the relationship between old B. pertussis isolates, vaccine seed isolates and recent isolates from Iran. B. bronchiseptica MO149 used as an outgroup. Isolated marked with black circle are vaccine seeds used in WCV manufactured by Serum Institute of India that is used for immunization in Iran. BP509 and BP134 were also used in WCV in the years when Iran manufactured and used its own vaccine.
Discussion
Iran has been using WCV for immunization against pertussis for more than 50 years and continues to use WCV with a high WCV coverage [Citation10]. It encountered an increase in pertussis cases since 2007 with highest number in 2013 with 1415 suspected cases per 100,000 [Citation15]. A previous PFGE study revealed that most isolates collected during 2009–2014 belonged to PFGE group 10 which is likely to be equivalent to the PFGE group SR11 in European countries [Citation40]. Furthermore, ACV antigen gene typing of a small number of isolates showed that ptxP3 isolates were predominant in Iran during 2008–2015 [Citation40,Citation41]. In this study we examined the genomic differences and relationships of ptxP3 isolates collected in Iran with WCV immunization and compared with ptxP3 isolates from countries with ACV vaccination. Our analysis of 55 Iranian B. pertussis isolates collected after the pertussis resurgence showed that Iranian ptxP3 isolates were grouped into 8 clades, each had separately derived from or closely related to isolates from other countries including USA, UK and Canada, suggesting multiple importation of B. pertussis into Iran and then diversification and expansion of different clades in parallel.
The results further confirmed that the ptxP3 strains have spread across the globe including WCV regions and can compete effectively against non-ptxP3 strains in a WCV immunised population. This study is one of the few evolutionary genetic studies of B. pertussis in the middle east region where most of its neighbouring countries are also using WCV and our study underscores the importance of surveillance of B. pertussis in these countries.
The expansion of ptxP3 strains has been suggested to be associated with the replacement of WCV by ACV although this hypothesis has been controversial [Citation42]. In Poland where WCV has also been in use for decades, in 2000–2013, ptxP1 isolates predominated with 77.5% [Citation43]. However, in a more recent study, 61.5% of the isolates during 2010–2016 were found to be ptxP3 and were attributed to the increasing use of ACV since 2013 for primary immunization [Citation43–45]. The near complete replacement of the B. pertussis population in Iran by ptxP3 strains where only WCV has been used further suggests that the expansion of ptxP3 strains was not driven by ACV selection alone. In China where ACV has replaced WCV in 2012, both ptxP3 and ptxP1 strains circulate with predominance of ptxP1. However, the ptxP1 strains have been found to be resistant to macrolides which are used for pertussis treatment and prophylaxis [Citation46]. Macrolide resistance may have acted as selection pressure to maintain the ptxP1 B. pertussis population in China [Citation46].
Comparative studies of ptxP3 and ptxP1 strains suggest that ptxP3 strains can outcompete ptxP1 strains in mice regardless whether the mice were immunised with ACV [Citation8] and proteomics studies suggest that ptxP3 strains may have become metabolically fitter [Citation47]. Several studies have shown that some genes involved in amino acid biosynthesis regulation and energy metabolism are differentially expressed in ptxP3 strains compared with ptxP1 strains [Citation5,Citation47,Citation48]. In this study 17 nsSNPs have been found in metabolic genes of which 14 were only in Iranian isolates. Three metabolic genes (BP1072 (ppk), BP2212 (fhp) and BP2405) have been reported to be upregulated in ptxP3 strains [Citation5], which suggests further potential adaptation. However, it is difficult to ascertain their effect as the mutations may be neutral or even mildly deleterious.
Continued adaptation by ptxP3 strains is evident in virulence associated genes [Citation5]. From 15 nsSNPs in 12 virulence associated genes found only in ptxP3 strains, eight were only in Iranian isolates (), of which three were in genes (bteA, sphB3, ompQ) which have been reported to be upregulated in ptxP3 strains [Citation5]. There were two novel SNPs in the bteA gene and in its promoter. bteA (BP0500) is known to be a BvgAS-regulated gene and thought to play a crucial role in T3SS-mediated cell death [Citation49]. It is also shown to be less sensitive to be regulated in ptxP3 isolates in the presence of sulphate [Citation50]. From six SNPs in ptxP3 isolates that were in genes encoding ACV antigens (fim3, fhaB, prn and ptxA, B and C), three were nsSNPs and all have been reported previously.
A prominent feature of B. pertussis evolution in ACV immunised host populations is the emergence and expansion of pertactin deficient strains [Citation4]. Prn deficient strains were first reported in France, and now in many countries including UK, Canada, US and Australia [Citation25,Citation26,Citation51–53]. Australia and US reported the highest percentages of Prn deficient isolates with 89.7% of the isolates from 2013 to 2017 in Australia and >85% in 2013 in the US [Citation26,Citation54]. Multiple mechanisms of Prn non-expression were found with the majority of the Prn deficiency due to damage of the prn gene by IS481 insertion at a hotspot in the prn structural gene [Citation54]. All known mechanisms of prn inactivation were examined from the genome sequence data of the Iranian isolates and none of the Iranian isolates was found to be potentially pertactin negative. Since WCV contained far more antigens than the ACVs that contain pertactin, there may be less selection pressure and less advantage for pertactin deficient B. pertussis mutants to emerge in a WCV immunized population.
The Iranian ptxP3 isolates were divided into multiple clades. Clades 5 and 8 are of particular interest. Clade 5 carried the prn9 allele exclusively. prn9 isolates were previously observed in Japan from 2008 to 2012 with low frequency [Citation55] and sporadic isolations in other countries [Citation56,Citation57]. However, it is unknown whether Japanese prn9 isolates were related to Iranian isolates or derived independently. It is possible that the increasing prevalence of prn9 isolates in the WCV population were due to the pressure of WCV. prn9 allele produces near the same amino acid sequence as prn2 with one additional GGFGP repeat in region 1 of the prn gene. A global comparison also showed that all prn9 isolates shared seven SNPs with a prn2 UK isolate UK98, suggesting that prn9 isolates may have arisen from a prn2 background. The additional repeat might affect epitope structure as difference in epitopes between Prn2 and Prn1 alleles were found [Citation58]. Clade 8 contained the highest number of isolates (20), mostly collected from Tehran (12/20). Clade 8 has 22 SNPs in one or more isolates that are located in genes or promoters reported to be upregulated in ptxP3 isolates [Citation5].
IR142 was closely related to B. pertussis strain 18322 that had diverged before the expansion of B. pertussis Tohama I lineage [Citation7]. This divergent lineage has rarely been isolated in recent years [Citation32, Citation34, Citation35]. Nevertheless, it is interesting to note that IR142 was isolated in 2015, suggesting that the lineage might still be circulating in Iran at a very low frequency [Citation34,Citation35].
Resurgence of pertussis in Iran may be associated with divergence between vaccine seeds and circulating strains. Iran used to administer the DTwP vaccine locally manufactured by Razi Serum Institute for decades [Citation59]. Due to reports of low immunogenicity and high adverse reactions of the vaccine [Citation39,Citation60], the local vaccine was replaced by other commercial vaccines in 2009, mostly the Indian DTwP. Furthermore, the increase in the pertussis incidence in Iran in recent years may indicate low efficiency of the vaccines used to control the disease [Citation11,Citation12]. It was suggested that bacterial strains or the vaccine formulation in some batches of vaccines might be the reason behind these issues [Citation59]. Comparison of the vaccine strains used in manufacturing the Iranian WCV (BP509 and BP134) and Indian WCV (BP509, BP134, BP25525 and BP6229) with currently circulating clinical isolates showed that none was close to the current predominant ptxP3 isolates in Iran (). BP509 that was used in both Iranian and Indian vaccines was more distinct compared to other vaccine seeds due to its allelic profile (ptxA4/prn7). BP134, also used in both Iranian and Indian vaccines was grouped with the now less predominant ptxP1/ptxA2 isolates in Iran. These data may explain why the local DTwP vaccine regardless of the quality variation between vaccine batches was less effective to control pertussis in Iran and substitution of vaccine seeds with a currently predominant strain for vaccine production may provide a more effective vaccine. However, more studies are needed to ascertain the contribution of the divergence between vaccine seeds and currently circulating strains to pertussis resurgence in Iran.
In summary, this study found that the Iranian B. pertussis isolates from a WCV immunised population carried mostly the ptxP3 allele similar to B. pertussis circulating in many countries where ACVs are used. The isolates were grouped into 8 clades which were independent importations from other countries and then local expansion. The Iranian ptxP3 B. pertussis population was not due to the emergence of a novel, hypervirulent clone or the expansion of a single lineage. These findings have important implications on the control of pertussis as current vaccines are not an effective measure to control the newly evolved ptxP3 strains. Further epidemiological surveillance is important to monitor the evolutionary changes of the B. pertussis population to help develop more effective strategies to control pertussis.
Supplemental Material
Download Zip (620.2 KB)Acknowledgments
F.S. and R.L. conceived and designed the research study; A.S. performed the experiments, analysed the data, and wrote the paper; S.O. analysed the data; V.J. and M.N. performed the bacterial isolation and confirmed the clinical specimens; S.M.Z handled the coordination between provinces; A.C.Y.T. and B.L. performed the sequencing of extracted DNA. All authors read and approved the final version of the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Burns DL, Meade BD, Messionnier NE. Pertussis resurgence: perspectives from the Working group Meeting on pertussis on the causes, possible paths forward, and gaps in our knowledge. J Infect Dis. 2014 Apr 1;209(Suppl 1):S32–S35. PubMed PMID: 24626870. doi: 10.1093/infdis/jit491
- Bouchez V, Guiso N. Bordetella pertussis, B. parapertussis, vaccines and cycles of whooping cough. Pathog Dis. 2015 Oct;73(7). PubMed PMID: 26242280. doi: 10.1093/femspd/ftv055
- Mooi FR, Van Der Maas NA, De Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation – two sides of the same coin. Epidemiol Infect. 2014 Apr;142(4):685–694. PubMed PMID: 23406868. doi: 10.1017/S0950268813000071
- Belcher T, Preston A. Bordetella pertussis evolution in the (functional) genomics era. Pathog Dis. 2015 Nov;73(8), ftv064. PubMed PMID: 26297914; PubMed Central PMCID: PMCPMC4626590. doi: 10.1093/femspd/ftv064
- King AJ, van der Lee S, Mohangoo A, et al. Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains. PLoS One. 2013;8(6):e66150. PubMed PMID: 23776625; PubMed Central PMCID: PMCPMC3679012. doi: 10.1371/journal.pone.0066150
- Xu Y, Liu B, Grondahl-Yli-Hannuksila K, et al. Whole-genome sequencing reveals the effect of vaccination on the evolution of Bordetella pertussis. Sci Rep. 2015 Aug 18;5:12888. PubMed PMID: 26283022; PubMed Central PMCID: PMCPMC4539551. doi: 10.1038/srep12888
- Bart MJ, Harris SR, Advani A, et al. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. MBio. 2014 Apr 22;5(2):e01074. PubMed PMID: 24757216; PubMed Central PMCID: PMCPMC3994516. doi: 10.1128/mBio.01074-14
- Safarchi A, Octavia S, Luu LD, et al. Better colonisation of newly emerged Bordetella pertussis in the co-infection mouse model study. Vaccine. 2016 Jul 25;34(34):3967–3971. PubMed PMID: 27346304. doi: 10.1016/j.vaccine.2016.06.052
- Moradi-Lakeh M, Esteghamati A. National immunization Program in Iran: whys and why nots. Hum Vaccin Immunother. 2013 Jan;9(1):112–114. PubMed PMID: 23442584; PubMed Central PMCID: PMCPMC3667923. doi: 10.4161/hv.22521
- Shahcheraghi F, Lotfi M N, Parzadeh M, et al. Isolation of Bordetella pertussis and Bordetella Parapertussis from clinical specimens at different provinces. J Mazandaran Univ Med Sci. 2012;22(88):2–8. eng.
- Saffar MJ, Ghorbani G, Hashemi A, et al. Pertussis resurgence in a highly vaccinated population, Mazandaran, north of Iran 2008-2011: an epidemiological analysis. Indian J Pediatr. 2014 Dec;81(12):1332–1336. PubMed PMID: 24788914. doi: 10.1007/s12098-014-1445-0
- Centre for Diseases Control and Prevention MoH, Islamic Republic of Iran. A comprehensive guidefor family physicians (2012): The contagious disease surveillance system Ministry of Health, Islamic Republic of Iran. Standard No.: ISBN:978-964-519-138-0.
- Shojaei J, Saffar M, Hashemi A, et al. Clinical and laboratory features of pertussis in hospitalized infants with confirmed versus probable pertussis cases. Ann Med Health Sci Res. 2014 Nov;4(6):910–914. PubMed PMID: 25506485; PubMed Central PMCID: PMCPMC4250990. doi: 10.4103/2141-9248.144911
- Khazaei S, Ayubi E, Mansori K, et al. Pertussis incidence by time, province and age group in Iran, 2006-2011. Iran J Public Health. 2016 Nov;45(11):1525–1527. PubMed PMID: 28028509; PubMed Central PMCID: PMCPMC5182268.
- The World Health Organization (WHO). World Health Statistics 2015. 2015.
- Centre for Diseases Control and Prevention MoH, Islamic Republic of Iran. A comprehensive guide for family physicians (2009): the contagious disease surveillance system Iran: Standard No.: ISBN:978-964-519-138-0.
- Safarchi A, Octavia S, Wu SZ, et al. Genomic dissection of Australian Bordetella pertussis isolates from the 2008–2012 epidemic. J Infect. 2016 Apr;72(4):468–477. PubMed PMID: 26826518. doi: 10.1016/j.jinf.2016.01.005
- Darling AE, Mau B, Perna NT. Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010 Jun 25;5(6):e11147. PubMed PMID: 20593022; PubMed Central PMCID: PMCPMC2892488. doi: 10.1371/journal.pone.0011147
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25(14):1754–1760. PubMed PMID: 19451168; PubMed Central PMCID: PMCPMC2705234. doi: 10.1093/bioinformatics/btp324
- Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009 Aug 15;25(16):2078–2079. PubMed PMID: 19505943; PubMed Central PMCID: PMCPMC2723002. doi: 10.1093/bioinformatics/btp352
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014 Jul 15;30(14):2068–2069. PubMed PMID: 24642063. doi: 10.1093/bioinformatics/btu153
- Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015 Nov 15;31(22):3691–3693. PubMed PMID: 26198102; PubMed Central PMCID: PMCPMC4817141. doi: 10.1093/bioinformatics/btv421
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012 May;19(5):455–477. PubMed PMID: 22506599; PubMed Central PMCID: PMCPMC3342519. doi: 10.1089/cmb.2012.0021
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011 Oct;28(10):2731–2739. PubMed PMID: 21546353; PubMed Central PMCID: PMCPMC3203626. doi: 10.1093/molbev/msr121
- Barkoff AM, Mertsola J, Pierard D, et al. Surveillance of circulating Bordetella pertussis strains in Europe during 1998 to 2015. J Clin Microbiol. 2018 May;56(5). doi:10.1128/JCM.01998-17. PubMed PMID: 29491017; PubMed Central PMCID: PMCPMC5925733.
- Weigand MR, Williams MM, Peng Y, et al. Genomic Survey of Bordetella pertussis Diversity, United States, 2000-2013. Emerg Infect Dis. 2019 Apr;25(4):780–783. PubMed PMID: 30882317; PubMed Central PMCID: PMCPMC6433035. doi: 10.3201/eid2504.180812
- Locht C, Geoffroy MC, Renauld G. Common accessory genes for the Bordetella pertussis filamentous hemagglutinin and fimbriae share sequence similarities with the papC and papD gene families. EMBO J. 1992 Sep;11(9):3175–3183. PubMed PMID: 1354611; PubMed Central PMCID: PMCPMC556850. doi: 10.1002/j.1460-2075.1992.tb05394.x
- Bouchez V, Hegerle N, Strati F, et al. New data on vaccine antigen deficient Bordetella pertussis isolates. Vaccines (Basel). 2015 Sep 14;3(3):751–770. doi:10.3390/vaccines3030751. PubMed PMID: 26389958; PubMed Central PMCID: PMCPMC4586476.
- Sealey KL, Harris SR, Fry NK, et al. Genomic analysis of isolates from the United Kingdom 2012 pertussis outbreak reveals that vaccine antigen genes are unusually fast evolving. J Infect Dis. 2015 Jul 15;212(2):294–301. PubMed PMID: 25489002. doi: 10.1093/infdis/jiu665
- Octavia S, Wu SZ, Kaur S, et al. Whole-genome sequencing and comparative genomic analysis of Bordetella pertussis isolates from the 2007-2008 epidemic in Israel. J Infect. 2017 Feb;74(2):204–207. PubMed PMID: 27914992. doi: 10.1016/j.jinf.2016.11.012
- Bowden KE, Weigand MR, Peng Y, et al. Genome structural diversity among 31 Bordetella pertussis isolates from two recent U.S. whooping cough Statewide Epidemics. mSphere. 2016 May-Jun;1(3). PubMed PMID: 27303739; PubMed Central PMCID: PMCPMC4888882. doi: 10.1128/mSphere.00036-16
- Park J, Zhang Y, Buboltz AM, et al. Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics. 2012 Oct 10;13:545. PubMed PMID: 23051057; PubMed Central PMCID: PMCPMC3533505. doi: 10.1186/1471-2164-13-545
- Kallonen T, Grondahl-Yli-Hannuksela K, Elomaa A, et al. Differences in the genomic content of isolates before and after introduction of pertussis vaccines in four European countries. Infect Genet Evol. 2011 Dec;11(8):2034–2042. PubMed PMID: 21964035. doi: 10.1016/j.meegid.2011.09.012
- Heikkinen E, Kallonen T, Saarinen L, et al. Comparative genomics of Bordetella pertussis reveals progressive gene loss in Finnish strains. PLoS One. 2007 Sep 19;2(9):e904. PubMed PMID: 17878939; PubMed Central PMCID: PMCPMC1975675. doi: 10.1371/journal.pone.0000904
- Cummings CA, Brinig MM, Lepp PW, et al. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J Bacteriol. 2004 Mar;186(5):1484–1492. PubMed PMID: 14973121; PubMed Central PMCID: PMCPMC344407. doi: 10.1128/JB.186.5.1484-1492.2004
- Zhang S, Xu Y, Zhou Z, et al. Complete genome sequence of Bordetella pertussis CS, a Chinese pertussis vaccine strain. J Bacteriol. 2011 Aug;193(15):4017–4018. PubMed PMID: 21622744; PubMed Central PMCID: PMCPMC3147532. doi: 10.1128/JB.05184-11
- Caro V, Bouchez V, Guiso N. Is the sequenced Bordetella pertussis strain Tohama I representative of the species? J Clin Microbiol. 2008 Jun;46(6):2125–2128. PubMed PMID: 18385436; PubMed Central PMCID: PMCPMC2446832. doi: 10.1128/JCM.02484-07
- Weigand MR, Peng Y, Loparev V, et al. Complete genome sequences of four Bordetella pertussis vaccine reference strains from Serum Institute of India. Genome Announc. 2016 Dec 22;4(6). PubMed PMID: 28007855; PubMed Central PMCID: PMCPMC5180383. doi: 10.1128/genomeA.01404-16
- Zarei S, Jeddi-Tehrani M, Akhondi MM, et al. Primary immunization with a triple diphtheria-tetanus-whole cell pertussis vaccine in Iranian infants: an analysis of antibody response. Iran J Allergy Asthma Immunol. 2009 Jun;8(2):85–93. PubMed PMID: 19671937.
- Sadeghpour Heravi F, Nikbin VS, Nakhost Lotfi M, et al. Strain variation and antigenic divergence among Bordetella pertussis circulating strains isolated from patients in Iran. Eur J Clin Microbiol Infect Dis. 2018 Oct;37(10):1893-1900. PubMed PMID: 30094521. doi: 10.1007/s10096-018-3323-6
- Sadeghpour Heravi F, Nikbin V, Shahcheraghi F. Allelic variations between vaccine strains and circulating strains in pxtP of Bordetella pertussis in Iran. Vaccine Res. 2015;2(3):19-23. eng. doi: 10.18869/acadpub.vacres.2.3.19
- Lan R, Octavia S. Vaccine-driven selection and the changing molecular epidemiology of Bordetella pertussis. In: Rohani P, Scarpino SV, editors. Pertussis: epidemiology, immunology, & evolution. Oxford: Oxford University Press; 2019. p. 166–181.
- Mosiej E, Zawadka M, Krysztopa-Grzybowska K, et al. Sequence variation in virulence-related genes of Bordetella pertussis isolates from Poland in the period 1959–2013. Eur J Clin Microbiol Infect Dis. 2015 Jan;34(1):147–152. doi:10.1007/s10096-014-2216-6. PubMed PMID: 25090968.
- Mosiej E, Krysztopa-Grzybowska K, Polak M, et al. Multi-locus variable-number tandem repeat analysis of Bordetella pertussis isolates circulating in Poland in the period 1959-2013. J Med Microbiol. 2017 Jun;66(6):753–761. PubMed PMID: 28598302. doi: 10.1099/jmm.0.000408
- Polak M, Zasada AA, Mosiej E, et al. Pertactin-deficient Bordetella pertussis isolates in Poland-a country with whole-cell pertussis primary vaccination. Microbes Infect. 2018 Dec 21. PubMed PMID: 30580013.
- Xu Z, Wang Z, Luan Y, et al. Genomic epidemiology of erythromycin-resistant Bordetella pertussis in China. Emerg Microbes Infect. 2019;8(1):461–470. PubMed PMID: 30898080; PubMed Central PMCID: PMCPMC6455148. doi: 10.1080/22221751.2019.1587315
- Luu LDW, Octavia S, Zhong L, et al. Comparison of the whole cell proteome and secretome of epidemic Bordetella pertussis strains from the 2008-2012 Australian epidemic under Sulfate-Modulating Conditions. Front Microbiol. 2018;9:2851. PubMed PMID: 30538686; PubMed Central PMCID: PMCPMC6277516. doi: 10.3389/fmicb.2018.02851
- Luu LDW, Octavia S, Zhong L, et al. Proteomic adaptation of Australian epidemic Bordetella pertussis. Proteomics. 2018 Apr;18(8):e1700237. PubMed PMID: 29464899. doi: 10.1002/pmic.201700237
- Han HJ, Kuwae A, Abe A, et al. Differential expression of type III effector BteA protein due to IS481 insertion in Bordetella pertussis. PLoS One. 2011 Mar 10;6(3):e17797. PubMed PMID: 21423776; PubMed Central PMCID: PMCPMC3053399. doi: 10.1371/journal.pone.0017797
- de Gouw D, Hermans PW, Bootsma HJ, et al. Differentially expressed genes in Bordetella pertussis strains belonging to a lineage which recently spread globally. PLoS One. 2014;9(1):e84523. PubMed PMID: 24416242; PubMed Central PMCID: PMCPMC3885589. doi: 10.1371/journal.pone.0084523
- Bouchez V, Brun D, Dore G, et al. Bordetella parapertussis isolates not expressing pertactin circulating in France. Clin Microbiol Infect. 2011 May;17(5):675–682. doi:10.1111/j.1469-0691.2010.03303.x. PubMed PMID: 20636430.
- Otsuka N, Han HJ, Toyoizumi-Ajisaka H, et al. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS One. 2012;7(2):e31985. PubMed PMID: 22348138; PubMed Central PMCID: PMCPMC3279416. doi: 10.1371/journal.pone.0031985
- Lam C, Octavia S, Ricafort L, et al. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis. 2014 Apr;20(4):626–633. PubMed PMID: 24655754; PubMed Central PMCID: PMCPMC3966384. doi: 10.3201/eid2004.131478
- Xu Z, Octavia S, Luu LDW, et al. Pertactin-negative and filamentous hemagglutinin-negative Bordetella pertussis, Australia, 2013-2017. Emerg Infect Dis. 2019 Jun;25(6):1196–1199. PubMed PMID: 31107218. doi: 10.3201/eid2506.180240
- Miyaji Y, Otsuka N, Toyoizumi-Ajisaka H, et al. Genetic analysis of Bordetella pertussis isolates from the 2008-2010 pertussis epidemic in Japan. PLoS One. 2013;8(10):e77165. PubMed PMID: 24124606; PubMed Central PMCID: PMCPMC3790747. doi: 10.1371/journal.pone.0077165
- Shuel M, Jamieson FB, Tang P, et al. Genetic analysis of Bordetella pertussis in Ontario, Canada reveals one predominant clone. Int J Infect Dis. 2013 Jun;17(6):e413–e417. PubMed PMID: 23352492. doi: 10.1016/j.ijid.2012.12.015
- Advani A, Donnelly D, Hallander H. Reference system for characterization of Bordetella pertussis pulsed-field gel electrophoresis profiles. J Clin Microbiol. 2004 Jul;42(7):2890–2897. PubMed PMID: 15243034; PubMed Central PMCID: PMCPMC446263. doi: 10.1128/JCM.42.7.2890-2897.2004
- He Q, Makinen J, Berbers G, et al. Bordetella pertussis protein pertactin induces type-specific antibodies: one possible explanation for the emergence of antigenic variants? J Infect Dis. 2003 Apr 15;187(8):1200–1205. PubMed PMID: 12695998. doi: 10.1086/368412
- Zarei S, Jeddi-Tehrani M, Akhondi MM, et al. Immunogenicity of a triple diphtheria-tetanus-whole cell pertussis vaccine in Iranian preschool children. Iran J Immunol. 2007 Jun;4(2):101–109. PubMed PMID: 17652850.
- Zarei S, Jeddi-Tehrani M, Mehdi Akhondi M, et al. Immunogenicity and reactogenicity of two diphtheria-tetanus-whole cell pertussis vaccines in Iranian pre-school children, a randomized controlled trial. Hum Vaccin Immunother. 2013 Jun;9(6):1316–1322. PubMed PMID: 23442608; PubMed Central PMCID: PMCPMC3901823. doi: 10.4161/hv.24093
