ABSTRACT
Molecular epidemiology of Clostridium difficile infection (CDI) has been extensively studied in North America and Europe; however, limited data on CDI are available in the Asia-Pacific region. A multicentre retrospective study was conducted in this region. C. difficile isolates were subjected to multilocus sequence typing (ST) and antimicrobial susceptibility testing. Totally, 394 isolates were collected from Hangzhou, Hong Kong, China; Busan, South Korea; Fukuoka, Japan; Singapore; Perth, Sydney, Australia; New York, the United States. C. difficile isolates included 337 toxin A-positive/B-positive/binary toxin-negative (A+B+CDT-), 48 A-B+CDT-, and nine A+B+CDT+. Distribution of dominant STs varied geographically with ST17 in Fukuoka (18.6%), Busan (56.0%), ST2 in Sydney (20.4%), Perth (25.8%). The antimicrobial resistance patterns were significantly different among the eight sites (χ2 = 325.64, p < 0.001). Five major clonal complexes correlated with unique antimicrobial resistances. Healthcare-associated (HA) CDI was mainly from older patients with more frequent antimicrobial use and higher A-B+ positive rates. Higher resistance to gatifloxacin, tetracycline, and erythromycin were observed in HA-CDI patients (χ2 = 4.76-7.89, p = 0.005-0.029). In conclusion, multiple C. difficile genotypes with varied antimicrobial resistance patterns have been circulating in the Asia-Pacific region. A-B+ isolates from older patients with prior antimicrobial use were correlated with HA-CDI.
Introduction
Clostridium difficile is a Gram-positive ubiquitous endospore-forming anaerobic bacterium that is a leading cause of antimicrobial-associated diarrhea and colitis [Citation1]. C. difficile infection (CDI) is a toxin-mediated disease, with clinical presentations ranging from mild self-limiting diarrhea to life-threatening pseudomembranous colitis, toxic megacolon, bowel perforation and sepsis [Citation2]. While CDI was initially mainly a healthcare-associated (HA) disease related to advanced age, exposure to antimicrobials and duration of hospitalization [Citation3–5], there has been an increase in community-associated (CA)- CDI worldwide [Citation6].
Recent global epidemics of CDI have shown that C. difficile is frequently transmitted between continents[Citation7,Citation8]. The epidemiology of CDI has been well documented in North America and Europe, particularly the emergence of ribotype (RT) 027 [Citation6,Citation9], however, the epidemiology continues to evolve. In comparison to these published in 2011 [Citation9], data from 2012 to 2014 in Europe showed that the distribution of the most common RTs has changed and new RTs have emerged with a wide diversity of genotypes in different countries[Citation10]. According to recent reports from the United States of America (USA) the proportion of RT 027 decreased significantly from 2011 to 2017 with increases of RTs: 106, 002, and 056 [Citation11].
With its large population and expanding economy over recent decades, Asia has been facing challenges related to many infectious diseases, including CDI [Citation12]. The mean overall prevalence of CDI in Asia was 14.8% with a pooled C. difficile incidence density of 5.3/10,000 patient days [Citation13]. A recent prospective study in Japan found a higher C. difficile incidence density (7.4/10,000 patient days) than that previously reported [Citation14]. Based on limited data from China, CDI among hospitalized patients with diarrhea had a pooled prevalence of 14.0% [Citation15]. However, in general, there is a paucity of data on CDI in Asia owing to poor awareness among clinicians and an underestimation of contribution of CDI to antimicrobial-related diseases [Citation12].
The molecular epidemiology of CDI in different regions in Asia is even more poorly understood than the traditional epidemiological features of the disease. There have been few studies in recent years with variable results among different countries. In China, C. difficile isolates with the dominant genotypes of ST2, ST3, ST35, and ST37 have been shown to be highly resistant to clindamycin and erythromycin, with high rates of multidrug resistance [Citation15–17]. In Japan, ST17, which includes the closely related RTs smz/RT018 and smz′/QX239, was the dominant genotype, followed by RT014/ST2, RT002/ST8, and RT369/ST81 [Citation12,Citation14]. ST17 was also the predominant genotype in South Korea, showing high resistance to clindamycin and moxifloxacin [Citation12,Citation18]. However, in general, the molecular characteristics and antimicrobial resistances of C. difficile strains circulating in patients with CDI in the Asia-Pacific region have not been well-described, and differences between HA- and CA-CDI in the Asia-Pacific region are still unclear.
Therefore, to improve our understanding of CDI epidemiology in the Asia-Pacific region, we conducted a multicenter retrospective study at seven clinical sites and one comparative clinical site to analyze differences in molecular features and antimicrobial resistance patterns of C. difficile isolates and investigate risk factors associated with HA- and CA-CDI.
Materials and methods
C. difficile isolates
In total, 394 C. difficile isolates were collected from CDI patients on a purely random basis without selection between January 2015 and March 2016. Isolates from Perth, Australia (n = 31); Sydney, Australia (n = 54); Busan, South Korea (n = 50); Singapore (n = 29); Fukuoka, Japan (n = 70); and Hong Kong, China (n = 50) were sent to our laboratory in transport medium for anaerobes. A total of 60 isolates were from New York, USA for the purpose of comparison. Fifty isolates were cultured from fecal samples collected from CDI patients in Hangzhou, China, as previous assays reported [Citation16]. All isolates were recovered on cefoxitin-cycloserine fructose agar plates (Oxoid, Basingstoke, United Kingdom) incubated for 48 h at 37°C in an anaerobic chamber with GENbag Anaer (bioMérieux, Marcy l’Etoile, France). Six control strains, including ATCC 43255 (RT087), ATCC BAA-1870 (ST1, RT027), and ATCC BAA-1801(RT010) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA).
Collection of clinical data
A standardized questionnaire was completed for each CDI patient at eight of the clinical sites, excluding Hong Kong and Singapore, to record sex, age, CDI type, CDI stage, antimicrobial treatment within the prior 8 weeks, and biochemical examinations, including white blood cells (WBCs), neutrophils, serum albumin, and creatinine, with cutoff values according to standard values [Citation19]. Patients with diarrhea induced by other pathogens were excluded and HA- and CA-CDI were defined on the basis of the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America guidelines [Citation3,Citation20]. Ethical approval for C. difficile isolate and clinical data collection was received from the Institutional Review Boards for each respective site. This study was approved by the Ethics Committee of Zhejiang Provincial Center for Disease Control and Prevention.
Detection of C. difficile toxin genes
Bacterial genomic DNA of C. difficile isolates and reference C. difficile strains (ATCC 43255, BAA-1870, and BAA-1801) were prepared using a DNeasy Blood & Tissue kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s protocol. The housekeeping gene tpi, toxin genes tcdA (toxin A) and tcdB (toxin B), and binary toxin genes cdtA and cdtB were detected by using a conventional PCR assay with the primer sequences as previously described [Citation21,Citation22]. The PCR amplicon sizes of cdtA and cdtB genes were 221-bp and 262-bp, respectively [Citation21]. The tcdA primers amplified a 369-bp amplicon for toxin A-positive/B-positive (A+B+) strains and a 110-bp amplicon for toxin A-negative/B-positive (A−B+) strains [Citation22]. After amplification, the PCR products were analyzed by agarose gel electrophoresis. A reference C. difficile strain (ATCC 43255) was used as a positive control for tcdA and tcdB and negative control for the binary toxin genes. C. difficile ATCC BAA-1870 was used as a positive control for the binary toxin genes, and C. difficile ATCC BAA-1801 was chosen as a negative control for tcdA and tcdB and the binary toxin genes. A blank, and positive and negative controls were examined in parallel for each test.
Multilocus sequence typing (MLST)
Seven housekeeping genes (adk, atpA, dxr, glyA, recA, sodA and tpi) were selected, and MLST was performed as previously described [Citation22]. PCR products were sequenced commercially by Sangon Biotech Co., Ltd. (Shanghai, China). Data for C. difficile alleles were deposited to determine STs in a public C. difficile MLST database (accessible at http://pubmlst.org/cdifficile).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by using the agar dilution assay described by the Clinical and Laboratory Standards Institute in 2017 [Citation23]. All isolates were subcultured on 5% Columbia blood agar (Oxoid, UK) and incubated for 48 h anaerobically at 37°C. After adjusting the turbidity to a 0.5 McFarland standard, aliquots (approximately 1 μL) of the cultures was spotted onto 1.2% Brucella agar (containing 5% lysed defiber sheep blood, 0.005‰ hemin, and 0.1% vitamin K1) plus antimicrobials at a given concentration using a multipoint inoculator and incubated anaerobically at 37°C for 48 h. The antimicrobials tested were as follows: piperacillin-tazobactam (PIP-TAZ), metronidazole, moxifloxacin, clindamycin, tetracycline, fusidic acid, vancomycin, rifampicin, erythromycin, ciprofloxacin, gatifloxacin, and levofloxacin. Interpretation of the minimal inhibitory concentration (MIC) results was based on previous studies [Citation16]. Recommended control strains of Bacteroides fragilis (ATCC 25285) and C. difficile (ATCC 700057) were utilized. The C. difficile isolates resistant to three antimicrobial classes was defined as multidrug resistance (MDR) according as previously described [Citation23].
Data analysis
Data were analyzed using Statistical Package for Social Sciences (SPSS, Chicago, IL, USA) version 22.0. Chi-square and Fisher’s exact tests were used to analyze correlations among clinical characteristics, toxin gene profiles, MLST types and antimicrobial susceptibility profiles of C. difficile isolates. The t-test and nonparametric statistics were used to compare differences in ages among all sites. P values were calculated to assess the differences among groups, and results with P values of less than 0.05 were considered statistically significant.
Results
Analysis of clinical data at six sites
Clinical information was obtained for 315 patients with CDI at six clinical sites, excluding Hong Kong and Singapore. All clinical data were analyzed and compared among different clinical sites (). The following parameters were found to be significantly correlated with the presence of CDI among all six sites: age, CDI type, fever higher than 38.3°C, neutrophils over 70%, serum albumin less than 35 g/L, creatinine more than 111 μM, and antimicrobial use within the prior 8 weeks (). No significant differences were found in WBCs, and the proportion of WBCs (cells × 109/L) of more than 10 at all sites was less than 50% (range: 26.7–44.3%). The mean age of patients with CDI was significantly younger in Hangzhou than in Busan (t = 5.19, P < 0.001), Fukuoka (Z = −5.18, P < 0.001), and New York (Z = −3.11, P < 0.001) and tended to be younger in Perth (t = 1.63, P = 0.107) and Sydney (Z = −1.67, P = 0.095). The proportions of HA-CDI were greater among older patients in Fukuoka and Hangzhou than in Busan (χ2 = 11.33, P = 0.001), New York (χ2 = 16.46, P < 0.001), Perth (χ2 = 18.55, P < 0.001), and Sydney (χ2 = 20.36, P < 0.001). The proportions of patients with CDI having a fever of higher than 38.3°C were significantly in Hangzhou and Fukuoka than in New York and Perth (χ2 = 13.93, P = 0.001). Only 8.6% of patients with CDI from Fukuoka had over 70% neutrophils, which was significantly lower than those from the other five sites (χ2 = 18.69–96.63, P < 0.001). The proportions of patients with CDI having serum albumin levels less than 35 g/L were much higher in Busan and Fukuoka than in the other four sites (χ2 = 33.49–86.32, P < 0.001). Furthermore, only 12.0% and 11.1% of patients with CDI had a history of antimicrobial treatment within the prior 8 weeks for cases occurring in Busan and Sydney; these values were much lower than those from other sites (χ2 = 21.61–91.67, P < 0.001).
Table 1. Clinical information of CDI patients.
Antimicrobial susceptibility analysis
The antimicrobial resistance patterns of 394 C. difficile isolates are presented in Table S1. Antimicrobial patterns were significantly different among eight clinical sites (χ2 = 325.64, P < 0.001). All isolates were susceptible to vancomycin and metronidazole, with MIC values of less than 0.5 and 2 μg/mL, respectively. No isolates were resistant to PIP-TAZ in Busan, New York, Singapore and Sydney. The rates of rifampin resistance were much lower than those of other antimicrobials in the Asia-Pacific region. The antimicrobial resistance rates were compared among three continents as follows. The rates of fusidic acid resistance in New York, Sydney, and Perth were significantly higher than those in sites from Asia (χ2 = 125.03, P < 0.001). However, the rates of moxifloxacin, tetracycline, and erythromycin resistance in C. difficile isolates from the three above sites were much lower than those from other sites (χ2 = 26.46–63.81, P < 0.001). The proportions of isolates resistant to gatifloxacin in two sites from Australia were significantly lower than those in other sites (χ2 = 52.84, P < 0.001). Notably, 186 (47.2%) of these isolates were MDR, although the rate of MDR was significantly lower in isolates from three sites in the USA and Australia than those from sites in Asia (χ2 = 51.35, P < 0.001).
Genotyping analysis of C. difficile isolates
Of the 394 C. difficile isolates, 337 (85.5%) were positive for both tcdA and tcdB and negative for both cdtA and cdtB (A+B+CDT−), 48 (12.2%) including one from New York, four from two sites in Australia, and 43 from other sites in Asia tested negative for tcdA and positive for tcdB without cdtA and cdtB (A−B+ CDT−), and nine (2.3%) including two from Australia, two from sites in Asia, and five from New York were positive for tcdA, tcdB, cdtA, and cdtB (A+B+CDT+).
The MLST analysis identified 68 different STs, and five C. difficile isolates were not typed because several housekeeping genes were not amplified. ST2 (n = 12, 4.8%), ST3 (n = 15, 6.0%), ST8 (n = 21, 8.4%), ST17 (n = 42, 16.9%), ST35 (n = 14, 5.6%), ST37 (n = 20, 8.0%), ST54 (n = 18, 7.2%), ST63 (n = 9, 3.6%), and ST81 (n = 17, 6.8%) were the dominant genotypes in Asia. However, the different distribution of STs was found in USA and Australia as follows. ST2 (n = 7, 11.7%) and ST42 (n = 11, 18.3%) were dominant STs in New York. ST2 (n = 19, 22.4%) and ST8 (n = 10, 11.8%) were dominant genotypes in two sites in Australia. There was a significant difference on the distribution of STs among three above regions (χ2 = 24.73, P < 0.001).
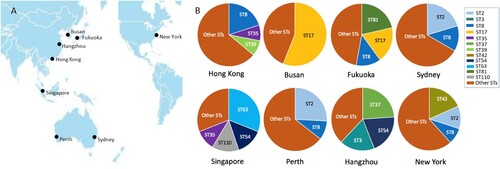
Different sites had a variety of STs, exhibiting varying distributions (). Two isolates from New York and Sydney were ST11. The CDT-positive isolates included three ST1 isolates (one from Hong Kong, two from New York), three ST41 isolates (one from New York, two from Perth), and three other STs (ST32 and ST67 from New York, ST95 from Singapore). The dominant genotypes between sites varied significantly, with ST17 in Fukuoka (n = 13, 18.6%) and Busan (n = 28, 56.0%); ST2 in New York as above, Sydney (n = 11, 20.4%) and Perth (n = 8, 25.8%); ST8 in Hong Kong (n = 10, 20.0%), Fukuoka (n = 9, 12.8%), and Sydney (n = 7, 13.0%); ST63 in Singapore (n = 9, 31.0%); ST42 in New York as above; and ST3 (n = 10, 20.0%), ST37 (n = 13, 26.0%), and ST54 (n = 10, 20.0%) in Hangzhou.
Figure 1. Distribution of STs at each site in this study. A: The seven sites are distributed in the Asia-Pacific region, and one comparative clinical site is New York, USA. B: Each site had various STs with different types of distribution. The major STs of C. difficile were labelled with different colours.
Comparison of clinical characteristics between HA- and CA-CDI
Of 315 patients with CDI with available clinical information, one patient from New York belonged to neither HA-CDI nor CA-CDI, exhibiting onset between 4 and 12 weeks after the final discharge, as previously published guidelines [Citation3,Citation20]. Clinical characteristics and genotypes were compared between HA- and CA-CDI for 255 patients in the Asia-Pacific region with retrievable C. difficile isolates (), the 59 patients from New York were also analyzed (data not shown). In comparison with CA-CDI, HA-CDI had a significantly higher frequency of age over 65 years and more frequent antimicrobial use. Moreover, patients with HA-CDI had a greater frequency of isolates with A−B+ genotype than patients with CA-CDI, although there were no significant differences in the distributions of STs between these groups (). Higher resistance rates to gatifloxacin, tetracycline, and erythromycin was observed in patients with HA-CDI (χ2 = 4.76–7.89, p = 0.005–0.029). C. difficile isolates related to CA-CDI had much higher rates of fusidic acid resistance than isolates related to HA-CDI in the Asia-Pacific region () and New York (data not shown). Furthermore, C. difficile isolates associated with CA-CDI had significantly higher rates of moxifloxacin resistance than isolates with HA-CDI in New York (Fisher’s exact test, p = 0.029) (data not shown).
Table 2. Differences of clinical characteristics, genotypes and antimicrobial resistance between HA-CDI and CA-CDI.
Phylogenetic analysis based on MLST
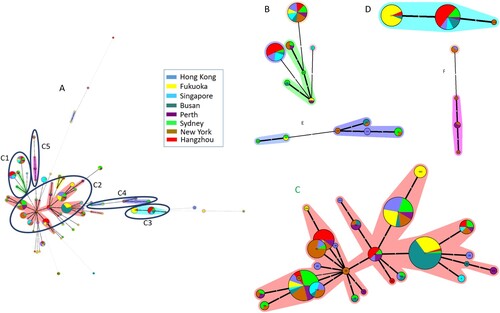
The genetic diversity and phylogenetic relationships of the 394 isolates were analyzed on the basis of MLST patterns. The minimum spanning tree (MST) was generated as shown in . In total, 68 STs were distributed in the phylogenetic tree. Five major clonal complexes (CC) were artificially divided based on the discriminating ability and typeability of MLST[Citation24,Citation25] according the certain assay, among which more than two allelic differences existed between each of the two clusters, and there were fewer than two allelic differences between STs in the same cluster. The detailed STs were shown in each CC in . The STs in CC1, CC2, and CC4 were distributed from various regions; all STs in CC3 belonged to A−B+ and mainly came from Hangzhou and Fukuoka (69.0%, 29/42); and STs in CC4 were predominantly from New York (62.5%, 5/8). The remaining STs were distributed outside of the five major CCs ().
Figure 2. Relationship of all C. difficile isolates based on MLST by MST. The number in the circle shows the ST type, and the size of circle corresponds the total number of C. difficile isolates belonging to this ST. The number of allelic differences between STs is showed on the branches. Nodes are connected by a dashed line if the allelic difference is over two alleles. Different colours correspond to different sites. A: The MST of C. difficile isolates in the study, B: A part of MST-CC1 including ST34, 35, 44, 54, 58, 95, and 357; C: A part of MST-CC2 including ST2, 3, 4, 8, 13, 14, 16, 17, 18, 28, 29, 32, 33, 42, 48, 49, 53, 66, 83, 92, 101, 102, 110, 183, 203, and 322; D: A part of MST-CC3 including ST37, 81, and 219; E: A part of MST-CC4 including ST43, 104, 129, 152, 235, and 239; F: A part of MST-CC5 including ST1, 41, 67, and 362.
Correlations between major genotypes and antimicrobial resistance patterns
Correlations between antimicrobial resistance and dominant genotypes are shown in Table S2. The antimicrobial patterns for A+B+ isolates were significantly different from those of A−B+ isolates (χ2 = 35.11, P < 0.001). Eleven PIP-TAZ-resistant isolates were distributed in different STs, and only one isolate was found in ST37 (A−B+). All ST81 isolates were susceptible to PIP-TAZ and rifampin, but presented higher resistance rates for other antimicrobials than those of other STs. ST37 isolates possessed a higher rifampin resistance rate than other STs (Fisher’s exact test, P < 0.001). ST2 isolates had lower resistance rates to moxifloxacin, tetracycline, and erythromycin than other antimicrobials. ST37 and ST17 isolates showed lower fusidic acid resistance rates than other STs, except ST3 and ST54 (χ2 = 10.35–53.25, P < 0.001). In general, the resistance patterns for most antimicrobials were significantly different among predominant STs (χ2 = 53.25–109.05, P < 0.001).
Correlations between major CCs and antimicrobial resistance patterns
The distributions of antimicrobial resistance patterns in five major CCs are shown in . The rates of resistance to fusidic acid, ciprofloxacin, and PIP-TAZ in CC3 were lower than those in other CCs, although there were no significant differences. However, C. difficile isolates had significantly higher rates of resistance to rifampin, fluoroquinolone, tetracycline, and erythromycin in CC3 in the A−B+ group than in other CCs in the A+B+ group (χ2 = 36.84–49.29, P < 0.001). No isolates resistant to PIP-TAZ, metronidazole, rifampin, moxifloxacin, vancomycin, tetracycline, and erythromycin were found in CC4, in which the rate of MDR was also significantly lower than in other CCs (Fisher’s exact test, P = 0.001).
Table 3. Difference of antimicrobial resistance patterns in five clonal complexes of C. difficile isolates.
Discussion
In recent years, an increasing number of studies has evaluated CDI epidemiology in Asia [Citation12,Citation26]. These have suggested that CDI is a very much under-recognized problem in Asia owing to poor clinical awareness among physicians [Citation12]. To date, small-scale data have described CDI epidemiology within different countries in Asia. However, the molecular characteristics and antimicrobial resistances of C. difficile isolates in the Asia-Pacific region have not been fully elucidated, despite the large population in this region. In this retrospective study, we found that there were significant differences in ST distributions and antimicrobial resistance patterns among the seven sites in the Asia-Pacific region and New York. Patients with HA-CDI tended to be older, have more frequent antimicrobial use, exhibit lower infection rates with A+B+ genotypes, and show higher resistance to gatifloxacin, tetracycline, and erythromycin than patients with CA-CDI in Asia-Pacific region but not in New York. Five major CCs correlated with unique antimicrobial resistances, and the rifampin, fluoroquinolone, tetracycline, and erythromycin resistance rates in the A−B+ cluster were significantly higher than those in the A+B+ group.
Interestingly, a different distribution of STs was found in different regions located in separate continents including Australia, USA, and Asian countries. Moreover, specific STs were dominant in neighbouring regions. ST17 was a dominant genotype in Fukuoka and Busan, both of which were located in Northeast Asia. Sydney and Perth, two major cities in Australia, had the same dominant genotype, ST2. Similar data has been reported previously [Citation12,Citation18]. In addition, different sites had their own dominant genotypes, i.e. ST3, ST37, and ST54 in Hangzhou; ST63 in Singapore; ST8 in Hong Kong; and ST42 in New York. Additionally, these sites showed different antimicrobial resistance patterns.
CDI epidemiology in America has been changing in recent years. ST42 has replaced ST1 as one of the most frequent types in the USA [Citation27], and ST42 has become one of the most common genotype in Brazil [Citation28]. However, CDI epidemiology in China presented stable, and ST3, ST37, and ST54 have been being dominant genotypes in different regions in China [Citation15,Citation16]. Consequently, our study demonstrated that multiple C. difficile genotypes are circulating in the Asia-Pacific region and exhibit unique molecular characteristics at each site. C. difficile isolates with the same STs may be transmitted between different regions through food trade and travel in the Asia-Pacific region as previously reported [Citation26]. Further studies focusing on analysis of whole-genome sequences combined with epidemiological data are required to investigate genetic relationship between different sites for the same STs.
There were statistically significant differences in antimicrobial resistance patterns not only among different sites but also among the major STs identified in our study. Our previous study also showed that the major genotypes were associated with significantly different antimicrobial resistance patterns in comparison with other genotypes in C. difficile in Eastern China [Citation16]. Antimicrobial resistance mechanisms in C. difficile have been investigated and associated with genes and gene mutations [Citation29]. Different STs obtained various mobile genetic elements containing different antimicrobial resistance-related genes at the different time in the course of genome evolution, maybe resulting in significant differences in antimicrobial resistance patterns among the STs [Citation8,Citation30]. Although extremely high rates of resistance to clindamycin, and levofloxacin were observed at all sites, New York, Sydney, and Perth showed lower rates for moxifloxacin, tetracycline, and erythromycin resistance than isolates from Asian locations. The low antimicrobial resistance rates in these sites could have several explanations. In Australia, the use of fluoroquinolones has historically been regulated and restricted in both humans and food-producing animals [Citation31]. Access to antimicrobials for clinical use can only be obtained through physician prescription and antimicrobials are not available for over the counter purchase. Furthermore, antimicrobial stewardship programmes were established during the early stages of epidemic outbreaks and strictly implemented in the USA and Australia, respectively [Citation32–35]. As inappropriate use of antimicrobials has been a major concern in recent years in Asia, a stewardship approach to antimicrobial use in humans and animals, when combined with enhanced infection control, may help to decrease antimicrobial resistance rates in the future.
Even though some clinical parameters were found to be significantly correlated with CDI, small numbers of samples might lead to possible bias in our data analysis results between sites. Thus, more clinical samples should be collected to investigate differences on clinical parameters of CDI patients among different countries in the Asia-Pacific region. Notably, we found that the mean age of patients with CDI in Hangzhou was younger than that in other sites despite small numbers of samples involved in this study. Similar results have been reported in the other studies of CDI epidemiology in China [Citation16,Citation36,Citation37]. Interestingly, we also found the high rates of HA-CDI with over 50% in each site, and HA-CDI tended to be older patients with more frequent antimicrobial use. Furthermore, C. difficile A−B+ genotypes infected more patients with HA-CDI, with higher rates of gatifloxacin, tetracycline, and erythromycin resistance, than patients with CA-CDI in the Asia-Pacific region. These above molecular characteristics of CDI were obviously different from those in New York. To the best of our knowledge, this was the first study elucidating the differences between patients with HA- and CA-CDI in the Asia-Pacific region and differences on molecular characteristics of CDI in between the Asia-Pacific region and the other region. While healthcare associated acquisition has been the primary pathway for CDI increasing incidence densities in the USA [Citation38], recent data indicated that over 40% of CDIs in the USA were community-associated [Citation6,Citation39]; in Europe, 76.4% of CDI cases were healthcare-associated [Citation40]. However, the main transmission pathway of CDI is still unknown in Asia, particularly China. Large-scale CDI epidemiological investigations are required to clarify the main route of transmission for CDI.
Our study had several limitations. First, all C. difficile isolates used in this study were not collected at the same period. CDI epidemiology is dynamically changing; thus, the distributions of genotypes and antimicrobial resistance patterns may have shown slight bias during data analysis. Second, this was a retrospective study involving relatively small numbers of cases recruited from each site. There were no detailed set inclusion or exclusion criteria thereby potentially resulting in a possible bias in our data analysis results between sites. Third, as there is a paucity of data on clinical outcomes in CDI patients, the differences of clinical outcomes among different regions and among CDIs with different molecular genotypes were not analyzed. Fourth, our study lacked some sites in Asian countries and did not cover the whole Asia-Pacific region. Thus, further investigations are required to clarify the differences of clinical outcomes induced by various genotypes of C. difficile in different regions, and confirm the molecular characteristics of C. difficile isolates in the Asia-Pacific region.
In conclusion, this was the first multicenter retrospective study in the Asia-Pacific region evaluating differences in the molecular features and antimicrobial resistance patterns of C. difficile isolates and investigating risk factors in patients with HA- and CA-CDI. Multiple C. difficile genotypes with varied antimicrobial resistance patterns have been circulating in the Asia-Pacific region. A−B+ isolates in older people and individuals with prior antimicrobial use were associated with HA-CDI. Consequently, CDI has a major presence in the Asia-Pacific region, and it is necessary to initiate CDI surveillance through international collaboration and implement antimicrobial stewardship programmes in order to prevent and control CDI.
Supplemental Material
Download MS Word (22.6 KB)Acknowledgements
We thank Wun-Ju Shieh for critically reviewing the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Martin JS, Monaghan TM, Wilcox MH. Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nat Rev Gastro & Hepatol. 2016 Apr;13(4):206–216. doi: 10.1038/nrgastro.2016.25
- Barbut F, Gariazzo B, Bonne L, et al. Clinical features of Clostridium difficile-associated infections and molecular characterization of strains: results of a retrospective study, 2000–2004. Infect Control Hosp Epidemiol. 2007 Feb;28(2):131–139. doi: 10.1086/511794
- Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol. 2010 May;31(5):431–455. doi: 10.1086/651706
- Iwashima Y, Nakamura A, Kato H, et al. A retrospective study of the epidemiology of Clostridium difficile infection at a University Hospital in Japan: genotypic features of the isolates and clinical characteristics of the patients. J Infect Chem. 2010 Oct;16(5):329–333. doi: 10.1007/s10156-010-0066-4
- Kim H, Jeong SH, Roh KH, et al. Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J Lab Med. 2010 Oct;30(5):491–497. doi: 10.3343/kjlm.2010.30.5.491
- Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. New Eng J Med. 2015 Feb 26;372(9):825–834. doi: 10.1056/NEJMoa1408913
- Cairns MD, Preston MD, Hall CL, et al. Comparative genome analysis and global phylogeny of the toxin variant Clostridium difficile PCR ribotype 017 reveals the evolution of two independent sublineages. J Clin Microbial. 2017 Mar;55(3):865–876. doi: 10.1128/JCM.01296-16
- Knight DR, Elliott B, Chang BJ, et al. Diversity and evolution in the genome of Clostridium difficile. Clin Microbial Rev. 2015 Jul;28(3):721–741. doi: 10.1128/CMR.00127-14
- Bauer MP, Notermans DW, van Benthem BH, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011 Jan 1;377(9759):63–73. doi: 10.1016/S0140-6736(10)61266-4
- Davies KA, Ashwin H, Longshaw CM, et al. Diversity of Clostridium difficile PCR ribotypes in Europe: results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Euro Surv. 2016 Jul 21;21(29). pii:30294.
- Tickler IA, Obradovich AE, Goering RV, et al. Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe. 2019 Jun 4. doi: 10.1016/j.anaerobe.2019.06.003
- Collins DA, Hawkey PM, Riley TV. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control. 2013 Jul 1;2(1):21. doi: 10.1186/2047-2994-2-21
- Borren NZ, Ghadermarzi S, Hutfless S, et al. The emergence of Clostridium difficile infection in Asia: a systematic review and meta-analysis of incidence and impact. PloS one. 2017;12(5):e0176797. doi: 10.1371/journal.pone.0176797
- Kato H, Senoh M, Honda H, et al. Clostridioides (Clostridium) difficile infection burden in Japan: a multicenter prospective study. Anaerobe. 2019 Mar 12;pii: S1075-9964(19):30046-0.
- Tang C, Cui L, Xu Y, et al. The incidence and drug resistance of Clostridium difficile infection in Mainland China: a systematic review and meta-analysis. Scientific Rep. 2016 Nov 29;6:37865. doi: 10.1038/srep37865
- Jin D, Luo Y, Huang C, et al. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in Eastern China. J Clin Microbial. 2017 Mar;55(3):801–810. doi: 10.1128/JCM.01898-16
- Chen YB, Gu SL, Wei ZQ, et al. Molecular epidemiology of Clostridium difficile in a tertiary hospital of China. J Med Microbial. 2014 Apr;63(Pt 4):562–569. doi: 10.1099/jmm.0.068668-0
- Seo MR, Kim J, Lee Y, et al. Prevalence, genetic relatedness and antibiotic resistance of hospital-acquired Clostridium difficile PCR ribotype 018 strains. Int J Antimicrob Agents. 2018 May;51(5):762–767. doi: 10.1016/j.ijantimicag.2018.01.025
- Ichihara K, Boyd JC. An appraisal of statistical procedures used in derivation of reference intervals. Clin Chem Lab Med. 2010 Nov;48(11):1537–1551. doi: 10.1515/CCLM.2010.319
- McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018 Mar 19;66(7):e1–e48. doi: 10.1093/cid/cix1085
- Persson S, Torpdahl M, Olsen KE. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008 Nov;14(11):1057–1064. doi: 10.1111/j.1469-0691.2008.02092.x
- Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010 Mar;48(3):770–778. doi: 10.1128/JCM.01796-09
- CLSI. Wayne (PA): Clinical and Laboratory Standards Institute.
- Killgore G, Thompson A, Johnson S, et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol. 2008 Feb;46(2):431–437. doi: 10.1128/JCM.01484-07
- Maiden MC, Jansen van Rensburg MJ, Bray JE, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013 Oct;11(10):728–736. doi: 10.1038/nrmicro3093
- Collins DA, Riley TV. Clostridium difficile in Asia: opportunities for one health management. Tropical Medicine and Infectious Disease. 2019;4(1):7. doi: 10.3390/tropicalmed4010007
- Kamboj M, Sheahan A, Sun J, et al. Transmission of Clostridium difficile during hospitalization for allogeneic stem cell transplant. Infect Control Hosp Epidemiol. 2016 Jan;37(1):8–15. doi: 10.1017/ice.2015.237
- Diniz AN, de Oliveira Junior CA, Vilela EG, et al. Molecular epidemiology of Clostridioides (previously Clostridium) difficile isolates from a university hospital in Minas Gerais, Brazil. Anaerobe. 2019 Apr;56:34–39. doi: 10.1016/j.anaerobe.2019.01.010
- Huang H, Weintraub A, Fang H, et al. Antimicrobial resistance in Clostridium difficile. Int J Antimicrob Agents. 2009 Dec;34(6):516–522. doi: 10.1016/j.ijantimicag.2009.09.012
- Dingle KE, Elliott B, Robinson E, et al. Evolutionary history of the Clostridium difficile pathogenicity locus. Gen Biol Evol. 2014 Jan;6(1):36–52. doi: 10.1093/gbe/evt204
- Cheng AC, Turnidge J, Collignon P, et al. Control of fluoroquinolone resistance through successful regulation, Australia. Emerg Infect Dis. 2012 Sep;18(9):1453–1460. doi: 10.3201/eid1809.111515
- Avent ML, Hall L, Davis L, et al. Antimicrobial stewardship activities: a survey of Queensland hospitals. Australian Heal Rev. 2014 Nov;38(5):557–563. doi: 10.1071/AH13137
- Fishman N. Policy statement on antimicrobial stewardship by the Society for Healthcare Epidemiology of America (SHEA), the Infectious Diseases Society of America (IDSA), and the Pediatric Infectious Diseases Society (PIDS). Infect Control Hosp Epidemiol. 2012 Apr;33(4):322–327. doi: 10.1086/665010
- Nagel JL, Kaye KS, LaPlante KL, et al. Antimicrobial stewardship for the infection control practitioner. Infect Dis Clin North America. 2016 Sep;30(3):771–784. doi: 10.1016/j.idc.2016.04.012
- Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis. 2016 May 15;62(10):e51–e77. doi: 10.1093/cid/ciw118
- Hawkey PM, Marriott C, Liu WE, et al. Molecular epidemiology of Clostridium difficile infection in a major Chinese hospital: an underrecognized problem in Asia? J Clin Microbiol. 2013 Oct;51(10):3308–3313. doi: 10.1128/JCM.00587-13
- Fang WJ, Jin DZ, Luo Y, et al. Clostridium difficile carriage in hospitalized cancer patients: a prospective investigation in eastern China. BMC Infect Dis. 2014 Sep 29;14:523. doi: 10.1186/1471-2334-14-523
- Magill SS, O’Leary E, Janelle SJ, et al. Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med. 2018 Nov 1;379(18):1732–1744. doi: 10.1056/NEJMoa1801550
- Dubberke ER, Olsen MA. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis. 2012 Aug;55(Suppl 2):S88–S92. doi: 10.1093/cid/cis335
- Krutova M, Kinross P, Barbut F, et al. How to: surveillance of Clostridium difficile infections. Clin Microbiol Infect. 2018 May;24(5):469–475. doi: 10.1016/j.cmi.2017.12.008
