?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Tigecycline is considered one of the last-resort antimicrobials for carbapenem-resistant K. pneumoniae. Plasmid-mediated tigecycline resistance remains largely unclear. Here, by utilizing whole genome sequencing, we report a plasmid-mediated tigecycline resistance mechanism, a 6,489 bp Resistance-nodulation-division family (RND) efflux pump (tmexCD1-toprJ1 pump), that confers transferable tigecycline resistance in K pneumoniae isolated from patients and chickens. In addition, we identified high prevalence of the plasmids co-harbouring both tmexCD1-toprJ1 pump and mcr (tmexCD1-mcr co-harbouring plasmid) from human in our nationwide collection. Even worse, the tmexCD1-toprJ1 and mcr co-harbouring plasmid was also co-existed with blaNDM-harbouring IncX3 plasmid in the same host, resulting in pandrug resistance. Phylogenetic analysis suggested that the plasmid-borne tmexCD1-toprJ1 originated from the chromosome of Aeromonas spp. through Tn5393-mediating translocation. Both plasmid-harbored tmexCD1-toprJ1 gene and mcr-8 likely originated from animal isolates and then spread to human. Our findings highlight a substantial threat of tmexCD1-toprJ1-mcr8 co-harbouring IncFIA/IncFII plasmid to public health due to their mobile resistance to both tigecycline and colistin, emphasizing an urgent need for further global surveillance on this plasmid.
Introduction
Carbapenem-resistant K. pneumoniae (CRKP), which account for 70–90% of clinical carbapenem-resistant Enterobacteriaceae (CRE) infections [Citation1, Citation2], poses a serious threat to global public health [Citation3, Citation4]. A number of studies have shown that infections caused by CRKP are associated greater disease severity, more comorbid conditions and increased mortality [Citation5, Citation6]. Tigecycline and colistin have been regarded as the “last resort” antimicrobials to fight against CRKP [Citation7, Citation8]. Unfortunately, the high prevalence of CRE has inevitably led to increased use of tigecycline and colistin, accelerating the emergence of tigecycline-resistant and colistin-resistant isolates. Remarkably, tigecycline resistance emerged soon after it was introduced into clinical practice in 2005. Chromosomal mutations, including overexpression of efflux pumps or mutations in the ribosome, have long been regarded as the major mechanisms leading to tigecycline-resistance in K. pneumoniae [Citation9]. Emergence and rapid global dissemination of colistin-resistant K. pneumoniae, which is due to plasmid-mediated colistin resistance (mcr) genes further worsened the situation [Citation10, Citation11]. In fact, plasmid-mediated resistance to several important antimicrobial classes, such as broad-spectrum beta-lactams and colistin have been found to facilitating resistant gene transfer from animals-derived Enterobacteriaceae to human-derived Enterobacteriaceae isolates [Citation12]. Of note, recent studies have shown that plasmid-mediated tigecycline resistance genes tet(X3), tet(X4), tet(X5) and tet(X6) are predominantly present in Enterobacteriaceae and Acinetobacter from human and animals [Citation13–20]. Interestingly, until now, only one tet(X3)-positive tigecycline-resistant K. pneumoniae were identified from a porcine caecum sample, and it remains unclear whether tet(X3) is present in human isolates. In the present study, we aimed to further investigate the mechanisms on plasmid-mediated tigecycline resistance by whole genome sequencing. In addition, we investigated whether plasmid-mediated tigecycline resistance co-existed with plasmid-mediated colistin resistance in the same isolate.
Materials and methods
Sample collection and bacterial strains
We collected five isolates of K. pneumoniae (KA1-KA5) with similar genetic background but substantial differences in resistance to tigecycline and colistin from chickens on the same farm in Shandong Province. Through whole-genome sequencing and bioinformatic analysis, we found the difference in tigecycline resistance was associated with a novel RND family pump, which we initially named as pmexAB-oprY. During submission of this manuscript, another group reported the same pump [Citation21]. To avoid any potential confusion in the future studies on this pump, we changed the name of this pump to tmexCD1-toprJ1, in consistent with Lv et al. [Citation21]. Then, we screened the tmexCD1-toprJ1 pump in isolates from both chickens and humans from several surveillances. The chicken-derived K. pneumoniae were from the animal surveillance from 2013 to 2016, which included a total of 125 K. pneumoniae isolated from animals from 4 farms in 3 provinces in China; and the human-derived K. pneumoniae were from CMSS (Chinese Meropenem Surveillance Study) and CRE network from 2010 to 2018, which included a total of 3047 K. pneumoniae isolated from inpatients from 27 provinces in China. The bacteria identification in this study was performed by MALDI-TOF MS (Bruker Daltonik GmbH, Bremen, Germany).
Patients information were not included in this study; thus, ethical approval was waived (Acceptance number of Ethics Review Committee of Peking University People's Hospital: 2016-PHB-135). All tigecycline-non-susceptible or colistin-non-susceptible isolates, which were determined as tigecycline MICs4 μg/ml or colistin MICs
4 μg/ml, were screened for tmexCD1-toprJ1 and mcr-8 by PCR, followed by Sanger sequencing. All of the primers used are listed in the appendix (Table S1). E coli J53 (sodium azide resistance) was used as the recipient strain in the conjugation experiment, and E. coli DH5α and K. pneumoniae ATCC 13883 were used as the recipient strains in the electroporation experiment.
The antimicrobial susceptibility testing
The MICs were determined by the agar dilution and broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. ATCC 25922 (Escherichia coli) and ATCC 27853 (Pseudomonas aeruginosa) served as quality control strains for susceptibility testing. The breakpoints of tigecycline and colistin for Enterobacteriaceae were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines and CLSI guidelines, respectively. Tigecycline epidemiological cut-off values (ECOFFs) for Enterobacteriaceae were set at >2 μg/ml for Enterobacteriaceae by EUCAST (https://eucast.org/clinical_breakpoints/), while colistin breakpoints for Enterobacteriaceae were set at >4 μg/ml by CLSI.
Conjugation
The transferability of tigecycline resistance genes was determined by the conjugation assay on the tigecycline resistance strains and azide-resistant E. coli J53 by filter mating. The donor and recipient strains were mixed at a ratio of 1:3 on a microporous membrane for 12 h. The transconjugants were then selected on China blue agar plates supplemented with either tigecycline (1 μg/ml) and azide (100 μg/ml), or with colistin (4 μg/ml) and azide (100 μg/ml). The transconjugants were confirmed by MALDI-TOF MS and PCR with specific primers (Table S1) and sequencing. Antimicrobial susceptibility was determined using the broth microdilution method.
Subcloning experiments
To confirm the role of tmexCD1-toprJ1 in tigecycline resistance, a 7,205 bp full-length ORF of tmexCD1-toprJ1 was amplified by PCR (Table S1). Subsequently, 0·3 pmol of purified PCR product was incubated with 1 μL T-Vector pMD-19 (TaKaRa) and 5 μL DNA Ligation Mighty Mix (TaKaRa) at 16°C overnight. The recombinant plasmid pUC19-tmexCD1-toprJ1 was then transformed into competent E. coli DH5α by incubating on ice for 30 min followed by incubating for 45 s at 42°C. Immediately thereafter, the DH5α was transferred to SOC medium (TaKaRa) at 37°C for one hour and then was coated on Luria–Bertani Agar (LB) medium containing 100 μg/ml of ampicillin at 37°C overnight. Visible strains after overnight were verified by PCR and then subjected to susceptibility testing using broth microdilution method. The recombinant vector was then extracted and transferred into K. pneumoniae ATCC 13883 via electroporation.
Acrb and ramA gene expression analysis using real-time reverse transcription PCR (RT–PCR)
The expression levels of acrAB efflux pump genes were analysed by RT–PCR. The primers for acrB, ramA and the rrsE were used as previously described [Citation22]. Briefly, bacteria were grown aerobically in LB broth until mid-log phase. DNase-treated RNA templates were prepared using the RNeasy Kit (Qiagen, Hilden, Germany). cDNA was generated from total RNA using random primer hexamers. RT–PCR was performed using a Roche 480 thermocycler with a SYBR green PCR master mix (TaKaRa). The PCR programme consisted of 5s at 95°C, followed by 40 cycles of 15 s at 95°C and 31 s at 58°C. Each sample was run in triplicate.
Growth rate assay
Bacteria were inoculated into 5 mL of Mueller-Hinton broth and incubated overnight at 37°C. Overnight cultures of E. coli J53 and the transconjugants were diluted 1:10000 in Mueller-Hinton broth, and growth curves were performed in triplicate by incubating the cultures for 24 h at 37°C. Bacterial growth was monitored by measuring the optical density of the culture at 620 nm by Varioskan Flash (Thermo Fisher ScientificTM, Shanghai, China).
Whole-genome sequencing
Total genomic DNA were extracted using the TIANamp Bacteria DNA Kit DP302 (Tiangen Biotech, Beijing, China) followed by genomic DNA sequencing with Illumina HiSeq X Ten platform, which produced 150-bp paired-end reads and at least 100-fold coverage of raw reads. 20 of the isolates were also extracted by the QIAGEN Large-Construct kit (Qiagen Sciences, Germantown, MD, USA) and sequenced by the PacBio RS II system (Pacific Biosciences) with a 10-kb size library and P6/C4 chemistry. The hybrid assembling was performed using Unicycler v0·4·6. All draft genome sequences were deposited into NCBI Genome database, and organized under BioProject ID PRJNA595047. All resistance genes were detected by the Resfinder (https://cge.cbs.dtu.dk/services/ResFinder/) and Basic Local Alignment Search Tool (BLAST).
Public genomic database searching
The tmexCD1-toprJ1 efflux pump and mcr-8 sequences were queried against NCBI RefSeq Assembly database. The meta information including collection date, organism, source and et al of all subject strains was collected. Any strains without year of collection were discarded.
Sequence annotation
The genome sequence was annotated by Rapid Annotation Subsequencing Technology (RAST) (http://rast.nmpdr.org/), Prokka v1·12 and BLAST, when applicable. Comparative analysis was performed by BLAST Ring Image Generator (BRIG), and genetic structures of representative plasmids were generated with DNAPlotter and Vector NTI. For Prokka, the assembly was first annotated by Prokka v1·12, and then combined with the best matches from blast hits (minimal identity 90%, minimal coverage 98%) against several specific databases, including SerotypeFinder, PlasmidFinder, ResFinder, CARD, VirulenceFinder, VFDB, ICEberg and ISFinder. Gene organizations were illustrated using R software.
Phylogenetic analysis and dating
Longest homology regions containing tmexCD1-toprJ1 or mcr-8 were determined based on pairwise Blast. Multiple sequence alignment of the homology regions was constructed by progressiveMauve, and all gaps were removed. Possible recombination events were detected by ClonalFrameML and removed. Phylogenetic tree was then constructed using BEAST under different site models, clock models and tree priors. Best model was selected based on BIC criteria. Divergence dating was inferred from MCMC trees. Phylogenetic tree was illustrated using R ggtree package.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
A novel efflux pump gene confers transferable resistance to tigecycline on plasmids
The five K. pneumoniae isolates (isolates KA1-KA5, ) from the same farm in Shandong displayed the same multilocus sequence typing (MLST) typing (ST37) but different levels of resistance to tigecycline. We screened major known chromosomal-mediated tigecycline resistance mechanisms, including ramA, ramR genes and acrAB efflux pump, but failed to find any differences between the resistant strains and susceptible strains. We then subjected all the five strains to whole genome sequencing including PacBio Sequencing. Cross-comparison of sequencing data among the five isolates revealed that the chromosome genomes of all isolates were highly similar (degree of matching over 99%) with slight difference in the plasmids. Further analysis of the plasmid sequences between tigecycline-non-susceptible K. pneumoniae (TNSKP) and tigecycline-susceptible K. pneumoniae (TSKP) from the five isolates motioned above highlighted a 6489 bp resistance-nodulation-cell division (RND) family efflux pump, which we suspected to be related to tigecycline resistance. The novel RND family efflux pump, which consists four ORFs (558, 1164, 3315, and 1434 bp in lengths, respectively), displayed the highest similarity to the mexCD efflux pump nfxB-mexC-mexD-oprJ from the mex family members with consistencies of 54%, 60%, 77% and 67%, respectively. We initially named this pump as pmexAB-oprY, which was changed to tmexCD1-toprJ1 in consistent with Lv et al [Citation21].
Table 1. Phenotype and genotype of 24 Klebsiella pneumoniae isolates that carrying tmexCD1-toprJ1 efflux pump or mcr-8 gene in this study.
The prevalence of tmexCD1-toprJ1 in TNSKPs
The sequencing data also revealed that four of 5 isolates (KA1, KA2, KA3 and KA5) contained mcr-8·1-harbouring plasmids, which led those isolates resistant to colistin. Most importantly, 2 isolates (KA1 and KA3) contained both mcr-8·1 and tmexCD1-toprJ1. We then did a retrospective screening on a large collection of isolates from our routine surveillance networks, including animal Network, CMSS and CRE Network. A total of 257 TNSKPs were obtained from 10536 Gram-negative bacilli, including 21 TNSKPs from animals and 236 TNSKPs from patients with various infections. Additional 8 isolates from chickens and 6 isolates from inpatients among the 257 TNSKPs were identified positive for the tmexCD1-toprJ1 gene. The prevalence of tmexCD1-toprJ1 in TNSKPs were 52·4% in animals and 2·5% in patients, respectively (table S3). Notably, all of tmexCD1-toprJ1-positive isolates were resistant to tigecycline, further reinforcing a link between tmexCD1-toprJ1 and tigecycline resistance.
Discovery of a new mcr-8 variant and the co-existing of multiple drug resistance genes in same isolates
Importantly, we noticed that among the 17 tmexCD1-toprJ1-positive isolates (14 isolates from the retrospective screening mentioned above and 3 isolates from the first 5 isolates that were positive for tmexCD1-toprJ1), 7 isolates (41·2%, 7/17) were positive for both mcr-8 (mcr-8·1, and mcr-8·5) and tmexCD1-toprJ1. In addition to mcr-8, mcr-3.11 was identified in one isolate (KA11). mcr-8·5 (GenBank accession number MN836537), a new mcr-8 variant found in this study, exhibited 6 single nucleotide mutation sites (C152T, G694T, T1462A, A1559T, C1578T, A1690G) compared to mcr-8·1, leading to changes in 5 amino acids (A51V, A232S, F488I, Y520F, N564D). The high prevalence (41·2%) of tmexCD1-toprJ1-mcr8-double positive isolates greatly attracted our attention. To understand the relationship between tmexCD1-toprJ1 and mcr-8 in the same strain, we selected 19 isolates (11 isolates from the 17 tmexCD1-toprJ1-positive isolates mentioned above and 8 isolates were mcr-positive but tmexCD1-toprJ1-negative strains with similar background with tmexCD1-toprJ1-positive isolates from our surveillance network), which were positive for tmexCD1-toprJ1 and/or mcr-8, for whole genome sequencing to investigate the underlying mechanisms of tigecycline resistance (). As shown in , the presence of tmexCD1-toprJ1 genes is associated with higher levels of tigecycline MICs, but not with colistin MICs or meropenem MICs. We also compared the expression levels of chromosomal-derived tigecycline resistance mechanism, and failed to detect any significant differences between tmexCD1-toprJ1-positive TNSKPs and TSKP, while tmexCD1-toprJ1-negative tigecycline-resistant K. pneumoniae displayed significantly higher expression levels of acrB and ramA compared to tmexCD1-toprJ1-positive tigecycline-resistant K. pneumoniae and TSKP (figure S1), suggesting that tigecycline resistance in tmexCD1-toprJ1-positive tigecycline-resistant K. pneumoniaes were not due to higher expression of those chromosomal-derived efflux pumps.
Figure 1. Genetic features of AMR genes and resistance phenotype. Upper panel is the MIC curves for different drugs. Lower panel is the genetic features of related AMR genes. Each dot represents the existence of the gene, and the known replication origin type is labelled at top-right. Linked dots by vertical line mean they are co-located on the same plasmid. The star after sample name mark the availability of PacBio sequencing.
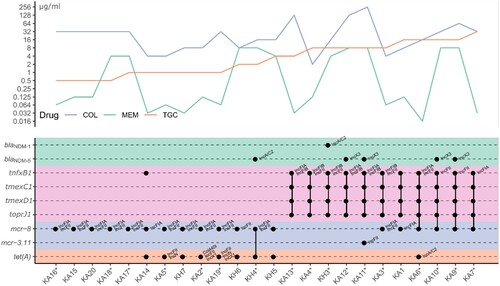
Even worse, we found that 40% (4/10) of the strains carrying both tmexCD1-toprJ1 and mcr were also positive for blaNDM (). Of particular clinical relevance is KH3, an isolate from urine sample from an inpatient in Beijing. KH3 was positive for tmexCD1-toprJ1, mcr-8·1 and blaNDM-1, making this strain resistant to all clinically available antibiotics ( and table S4).
One plasmid carries both mcr and tmexCD1-toprJ1 in 4 isolates from chickens
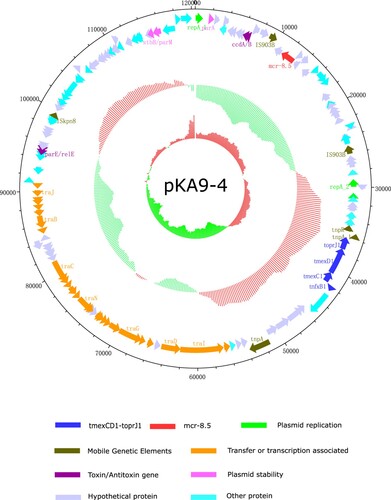
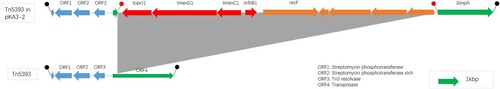
Remarkably, further comparative analysis of PacBio and HiSeq sequencing data revealed co-existence of mcr-8 and tmexCD1-toprJ1 in the same plasmid in four isolates (KA6, KA7, KA9 and KA10). Specifically, mcr-8·1 co-existed with tmexCD1-toprJ1 on IncFIA plasmid in two isolates (KA7 and KA9), while mcr-8·5 co-existed with tmexCD1-toprJ1 on IncFII(K) plasmid in the other two isolates (KA6 and KA10) (). For tmexCD1-toprJ1-harbouring plasmid, IncFIA was the most popular type, accounting for 45·5% (5/11) of tmexCD1-toprJ1-positive isolates, followed by IncFII (27·3%, 3/11) and IncFIB (27·3%, 3/11). Interestingly, all mcr-8·1 was located in IncFIA, while mcr-8·5 was located in IncFII(K) plasmid (). pKA9-4 (GenBank accession number MN832595), a 120-kb IncFII(K) plasmid, displayed 74% query coverage and 97·02% identity to the mcr-8-harbouring plasmid pKP91 from K. pneumoniae (Genbank accession number, MG736312) by Blast in the NCBI database (). In addition, further analysis showed that all the tmexCD1-toprJ1 efflux pumps were embedded in transposon Tn5393 (). Taken together, these findings demonstrated that the high prevalence of tmexCD1-toprJ1-mcr8-double positive K. pneumoniae is partially due to dissemination of plasmids that carry both mcr-8 and tmexCD1-toprJ1.
Function of tmexCD1-toprJ1 and the transferability of the plasmid carrying the tmexCD1-toprJ1
To directly confirm the function of the tmexCD1-toprJ1 pump, we performed a series of conjugation assays on KA1, KA3 and other three K. pneumoniae isolates (KA6, KA7 and KA9), which contained both mcr-8·1 and tmexCD1-toprJ1. Although the conjugation assays on KA1 and KA3 failed, the conjugation assay on other three isolates succeeded. As expected, the plasmid carrying the tmexCD1-toprJ1 pump greatly increased tigecycline resistance in E· coli J53 by at least 8-fold with transfer frequencies of 1·20±0·326 × 10−7. We next ligated the tmexCD1-toprJ1 gene to vector pUC-19 and transferred the plasmid pUC-19-tmexCD1-toprJ1 into E· coli DH5α and K. pneumoniae ATCC 13883. The antimicrobial susceptibility results showed that pUC-19-tmexCD1-toprJ1 increased the tigecycline MIC in DH5α by 8-fold and in K. pneumoniae ATCC 13883 by 16-fold, respectively (). In addition, the reduced susceptibility was also observed for tetracycline and ciprofloxacin ( and table S2). Taken together, these results show that tmexCD1-toprJ1 confers mobile tigecycline resistance in the host. Further, the effect of acquiring tmexCD1-toprJ1-and/or mcr-8-harbouring plasmids on biological fitness cost was evaluated. No significant differences in the growth rates were observed among the recipient strain J53, the mcr-8-plasmid-transconjugants strain KH3-T and the tmexCD1-mcr-8-co-harbouring plasmid plasmid-transconjugants KA6-T (Figure S2). These results indicate that acquiring plasmids co-carrying tmexCD1-toprJ1 and mcr-8 does not have a significant effect on fitness cost.
Table 2. Antimicrobial susceptibility profiles of the isolates carrying the tmexCD1-toprJ1 and their transconjugants and transformants.
Origin and evolution of tmexCD1-toprJ1 and mcr-8
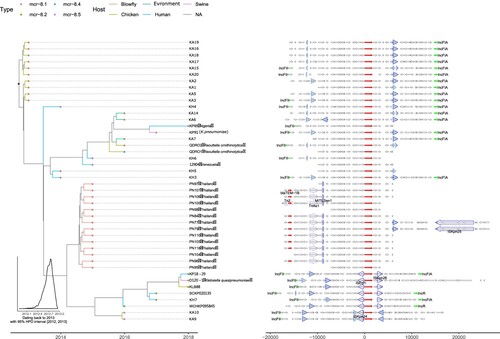
To gain insight into the spatiotemporal origin of the tmexCD1-toprJ1 gene, NCBI genomic database search was performed for strains containing the tmexCD1-toprJ1 gene. A total of 15 strains were found based on the criteria that the uploaded sequence from each strain was greater than 20 kb and had 100% coverage of tmexCD1-toprJ1 gene. Of note, phylogenetic tree and dating were performed based on the tmexCD1-toprJ1 sequences from 11 TNSKPs in this study and the 15 strains from the NCBI (). Interestingly, the tmexCD1-toprJ1 gene was located on the chromosome in almost all non-K. pneumoniae organisms. In contrast, the tmexCD1-toprJ1 gene was found inside Tn5393 on plasmid in all K. pneumoniae (). Importantly, the sequence contexts from Aeromonas (WCHAH045096 and WCW1-2) were found more similar to those seen in K. pneumoniae than Pseudomonas (). Thus, it is likely that the plasmid-harbored tmexCD1-toprJ1 gene originated from the chromosome of Aeromonas spp. through Tn5393 mediated translocation. Of note, there were at least one type of IncFIA plasmid and two types of IncFII plasmids harbouring Tn5393::tmexCD1-toprJ1 (tmexCD1-toprJ1 inserted in Tn5393), indicating this transposon may still possess the translocation capacity. The transposon in IncFIA plasmids (KA11, KA12 and KA13) were truncated from first three ORFs by IS1X4 and IS26, while the transposon in one IncFII plasmid (KP18-29) lost the transposase gene due to the replacement of tandem of insertion sequences. In addition, two strains (KP18-29 and KH3) isolated from human were found being surrounded by chicken-derived strains in the phylogenetic tree, suggesting a possibility of chicken to human transmission, although we cannot exclude the possibility of human to chicken transmission.
Figure 4. Phylogenetic tree and sequence context comparison of tmexCD1-toprJ1. Left panel is the phylogenetic tree inferred by BEAST2. The fill colours of tip nodes represent the sample source, and the line colours of tip branches demonstrate the hosts of strains. The insert histogram is the distribution of dating year for the root node. Right panel shows the sequence context. Thin arrows are genes, and fat arrows are insertion sequences where the dashed lines marks truncated ones. The highlighted (blue filled) insertion sequences are Tn5393.

The spatiotemporal origin of a new mcr-8 variant mcr-8·5 was assessed. Phylogenetic analysis and gene context comparison revealed that mcr-8·5 clustered with mcr-8·2 and they shared larger amount of sequence context. Thus mcr-8·5 was more likely derived from mcr-8·2. It is noticeable that, in comparison to mcr-8.1, mcr-8·2 and mcr-8·5 were surrounded by complete insertion sequences (ISKpn26, ISEcl1 and IS903B), which makes them to have higher risk for transposition and dissemination. In addition, we dated the emergence of mcr-8 back to 2013 (). Of note, as the chicken-derived strains were found quite close to that origin time, it is likely that mcr-8 was originated from animal and subsequently spread to human.
Figure 5. Phylogenetic tree and sequence context comparison of mcr-8. Left panel is the phylogenetic tree inferred by BEAST2. The fill colours of tip nodes represent different subtypes of mcr-8, and the line colours of tip branches demonstrate the hosts of strains. The insert histogram is the distribution of dating year for the root node. Right panel shows the sequence context. Thin arrows are genes, and fat arrows are insertion sequences where the dashed lines marks truncated ones. The highlighted (blue filled) insertion sequences are IS903B.
Discussion
Conjugative plasmids play an essential role in the dissemination of antimicrobial resistance among clinically important pathogens, such as K. pneumoniae. In the present study, we identified a plasmid-mediated tigecycline resistance mechanism, mediating by the tmexCD1-toprJ1 pump, in TNSKPs isolated from both patients and chickens. We also found the presence of plasmids co-harbouring tmexCD1-toprJ1 and mcr-8.5 genes, which allows simultaneous transmission of tigecycline resistance and colistin resistance in a single transfer event. Given previous experience on the rapid dissemination of carbapenem-resistant plasmids (blaKPC-2 or blaNDM-1 plasmids) and colistin-resistant plasmid (mcr plasmids), we highly suspect that the emergence of transmissible tigecycline-resistant plasmids (tmexCD1-toprJ1 plasmids), especially the emergence of plasmid co-harbouring both tmexCD1-toprJ1 and mcr, will largely promote the progression of global pan-drug resistance.
The novel plasmid-harbouring tmexCD1-toprJ1 pump belonged to a mex family pumps. The mex family proteins were first reported in the chromosomal genome of Pseudomonas aeruginosa as efflux pumps to confers P. aeruginosa intrinsic resistance to tetracycline, chloramphenicol and norfloxacin [Citation23]. Interestingly, although the mex family genes have been extensive studies in P. aeruginosa, in terms of plasmid-harbored tmexCD1-toprJ1 gene, it is more likely originated from the chromosome of Aeromonas spp. as assessed by the sequence contexts.
The recent emergence of plasmid-mediated tigecycline resistance genes tet(X3) and tet(X4) have attracted intense attention [Citation16]. Interestingly, until now, tet(X3) and tet(X4) were predominantly found in E. coli, and have never been reported in tigecycline-resistant K. pneumoniae isolated from human origin. In fact, we have screened a total of 161 TNSKPs from patients with various infections and failed to detect any tet(X3)/tet(X4)-positive TNSKPs (data not shown). In contrast, we found that plasmid-harbored tmexCD1-toprJ1 gene was present in TNSKP from human origin with a prevalence of 2·5%. In search of the public database, we found a human-derived K. pneumoniae plasmid contig containing the same sequence as tmexCD1-toprJ1 from Henan province (China) in 2018, which has been recently submitted to the NCBI (GenBank accession number MK262712·1) but was not annotated. Although no detailed information is available, the data confirms the presence of tmexCD1-toprJ1 in K. pneumoniae from human origin. Just recently, Lv et al. also found this pump in isolates from human [Citation21]. In our study, tmexCD1-toprJ1 was first identified in TNSKPs from inpatients in 2012 from two Provinces in China (Jiangsu and Tianjin), and then was found in TNSKP from an inpatient in Beijing in 2014. Of interest, tmexCD1-toprJ1 was re-emerged in 2018 from an inpatient in Xinjiang province. These data indicate that tmexCD1-toprJ1-positive TNSKP have been persistently present in human.
It should be noted that in vitro transfer of tmexCD1-toprJ1 only led to moderate tigecycline MIC increase by 8–16-fold, which was lower compared to that in tet(X) (64–128-fold). However, due to the efflux pump nature of tmexCD1-toprJ1, it not only mediates tigecycline resistance, but also may mediate resistance to other antimicrobials, such as ciprofloxacin, tetracycline and chloramphenicol.
It was surprising to find the high prevalence of tmexCD1-toprJ1 in TNSKPs (52·4%) from the animal origins. It is likely that the tmexCD1-toprJ1 had already disseminated in animals in China. Given the sharp differences in the prevalence of tmexCD1-toprJ1-positive TNSKP between animal origin and human origin (52·4% vs. 2·5%) as well as the phylogenetic findings, we suspect that tmexCD1-toprJ1-mediated tigecycline resistance originated in chickens and subsequently spread to human through plasmid-mediated gene transfer. Interestingly, tigecycline has never been introduced to veterinary practice, but other tetracyclines, such as oxytetracycline and doxycycline were heavily used in chickens [Citation24]. It is likely that continuous abuse of those tetracyclines impose continuous selective pressure to facilitate the spread of tmexCD1-toprJ1 gene in animal-derived isolates. Thus, it is of paramount important for multi-center surveillance and molecular epidemiological studies on the distribution and dissemination of tmexCD1-toprJ1 in both veterinary side and clinical side.
The co-existence of tmexCD1-toprJ1 with mcr-8 in the same plasmid deserves much attention, as our conjugation assay and phylogenic analysis reveal a high transferability of the tmexCD1-toprJ1-mcr-8 co-harbouring plasmid. Given this, further acquisition of carbapenem resistance genes by strains with tmexCD1-toprJ1-mcr-8 co-harbouring plasmid, or vice versa, would inevitably lead to pan-drug-resistant strains. In fact, we found that the tmexCD1-toprJ1-mcr-8 co-harbouring plasmid was indeed co-existed with blaNDM-harbouring plasmid in two strains. It is notable that one of the major tmexCD1-toprJ1-mcr-8 co-harbouring plasmid types is IncFII plasmid. Previous studies from our groups and others have demonstrated that the major carbapenem resistance genes blaKPC-harbouring plasmid type is also IncFII plasmid [Citation25]. Given the high prevalence of blaKPC-CRKP in China, it is highly likely that tmexCD1-toprJ1-mcr-8 co-harbouring plasmid acquires blaKPC, making a “super-drug resistant” plasmid.
A new mcr-8 variant mcr-8·5 was identified in two isolates. mcr-8 was recently reported in a CRPK from the pig in China [Citation26]. Subsequently, mcr-8 variants mcr-8·2 and mcr-8·3 were characterized in K. pneumoniae and mcr-8·4 were found in Raoultella ornithinolytica [Citation27, Citation28]. mcr-1 was the first identified plasmid-mediated colistin-resistant gene, has gained extensive attention [Citation11]. Interestingly, in the case of co-existence with tmexCD1-toprJ1, only mcr-8 and its variant mcr-8·5, as well as mcr-3·11 were identified. Of note, mcr-1 and mcr-8 display distinct preference to plasmids (i.e. mcr-1 prefers IncI2 and IncX4, while mcr-8 prefers IncFI and IncFII), while mcr-8 displays similar plasmid preference to tmexCD1-toprJ1 (i.e. both mcr-8 and tmexCD1-toprJ1 prefer IncFI and IncFII), it is likely that the common preference to the same plasmid determines the co-existence of mcr-8 with tmexCD1-toprJ1.
Similar to tmexCD1-toprJ1, phylogenetic analysis also reveals that mcr-8·5 was transmitted from animal to human. Colistin had been used as an animal feed additive in China for a long time, leading to the emergence, expansion and widespread of mcr-bearing plasmids. Fortunately, the Chinese Ministry of Agriculture announced a ban on use of colistin in animals in 2017 [Citation29]. With the introduction of polymyxin B (Approved in Jan. 2018) and colistin (Approved in Jul. 2019) into clinical practice, active surveillance of those mcr-bearing plasmids is of great importance.
In summary, our study deserves much more global attention, as our data clearly point out that tmexCD1-toprJ1-mediated tigecycline-resistance mechanism is already present in isolates from patients. Even worse, we show that tmexCD1-toprJ1-mcr-8 co-harbouring plasmid co-existed with blaNDM-harbouring plasmid in a single isolate, which is a truly “super-drug resistant bug,” already caused infections in patients. Thus, more effort is needed to elucidate the risk factors and clinical outcomes of patients with this “superbug.” Given that both tmexCD1-toprJ1 and mcr-8·5 are more likely to originate from animal, our data also emphasizes the urgent need for a “one-health” strategy and the global epidemiological surveillance landscape for the tmexCD1-toprJ1-harbouring plasmid, especially the tmexCD1-toprJ1-mcr-8 co-harbouring plasmid.
Supplemental Material
Download ()Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number 81625014). The authors thank Prof. Yuqing Liu (Shandong Academy of Agricultural Sciences) for collecting partial isolates. The authors also thank all the partners in the CMSS (Chinese Meropenem Surveillance Study) and CRE network for their contribution to this study. HW conceived the project and designed the experiments. SS and HG performed the experiments. SS, HG, RW, LJ, XW, QW, YY and HC collected the data. SS, HG, YL, RW, LJ and HW analysed and interpreted the data. SS, HG, YL and HW wrote the report. All authors reviewed, revised, and approved the final report.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017 May;19:98–106. doi:10.1016/j.ebiom.2017.04.032. PubMed PMID: 28479289; PubMed Central PMCID: PMCPMC5440625. doi: 10.1016/j.ebiom.2017.04.032
- Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17(2):153–163. doi:10.1016/s1473-3099(16)30257-2.
- Tzouvelekis LS, Markogiannakis A, Psichogiou M, et al. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012 Oct;25(4):682–707. doi:10.1128/CMR.05035-11. PubMed PMID: 23034326; PubMed Central PMCID: PMCPMC3485753. doi: 10.1128/CMR.05035-11
- Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/s1473-3099(17)30489-9.
- Cienfuegos-Gallet AV, de Los Rios AM O, Viana P S, et al. Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: a case-control and cohort study. BMC Infect Dis. 2019 Oct 7;19(1):830), doi:10.1186/s12879-019-4461-x. PubMed PMID: 31590648; PubMed Central PMCID: PMCPMC6781339.
- Martin A, Fahrbach K, Zhao Q, et al. Association between carbapenem resistance and mortality among adult, hospitalized patients with serious infections due to Enterobacteriaceae: results of a systematic literature review and meta-analysis. Open Forum Infect Dis. 2018 Jul;5(7):150. doi:10.1093/ofid/ofy150. PubMed PMID: 30046639; PubMed Central PMCID: PMCPMC6054228.
- Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017 Apr;30(2):557–596. doi:10.1128/CMR.00064-16. PubMed PMID: 28275006; PubMed Central PMCID: PMCPMC5355641. doi: 10.1128/CMR.00064-16
- Seifert H, Blondeau J, Dowzicky MJ. In vitro activity of tigecycline and comparators (2014-2016) among key WHO ‘priority pathogens’ and longitudinal assessment (2004-2016) of antimicrobial resistance: a report from the T.E.S.T. study. Int J Antimicrob Agents. 2018 Oct;52(4):474–484. doi:10.1016/j.ijantimicag.2018.07.003. PubMed PMID: 30012439. doi: 10.1016/j.ijantimicag.2018.07.003
- Chen Y, Hu D, Zhang Q, et al. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front Cell Infect Microbiol. 2017;7:37. doi:10.3389/fcimb.2017.00037. PubMed PMID: 28261566; PubMed Central PMCID: PMCPMC5313504.
- Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/s1473-3099(15)00424-7.
- Wang R, van Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018 Mar 21;9(1):1179, doi:10.1038/s41467-018-03205-z. PubMed PMID: 29563494; PubMed Central PMCID: PMCPMC5862964. doi: 10.1038/s41467-018-03205-z
- Dolejska M, Papagiannitsis CC. Plasmid-mediated resistance is going wild. Plasmid. 2018 Sep;99:99–111. doi:10.1016/j.plasmid.2018.09.010. PubMed PMID: 30243983. doi: 10.1016/j.plasmid.2018.09.010
- Sun J, Chen C, Cui CY, et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol. 2019 Sep;4(9):1457–1464. doi:10.1038/s41564-019-0496-4. PubMed PMID: 31235960; PubMed Central PMCID: PMCPMC6707864. doi: 10.1038/s41564-019-0496-4
- He T, Wang R, Liu D, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019 Sep;4(9):1450–1456. doi:10.1038/s41564-019-0445-2. PubMed PMID: 31133751. doi: 10.1038/s41564-019-0445-2
- Chen C, Cui CY, Zhang Y, et al. Emergence of mobile tigecycline resistance mechanism in Escherichia coli strains from migratory birds in China. Emerg Microbes Infect. 2019;8(1):1219–1222. doi:10.1080/22221751.2019.1653795. PubMed PMID: 31429665; PubMed Central PMCID: PMCPMC6713155. doi: 10.1080/22221751.2019.1653795
- Bai L, Du P, Du Y, et al. Detection of plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong provinces, China. Euro Surveill 2019 Jun;24(25). doi:10.2807/1560-7917.ES.2019.24.25.1900340. PubMed PMID: 31241040; PubMed Central PMCID: PMCPMC6593906.
- Chen C, Wu XT, He Q, et al. Complete sequence of a tet(X4)-harboring IncX1 plasmid, pYY76-1-2, in Escherichia coli from a cattle sample in China. Antimicrob Agents Chemother. 2019 Oct 7. doi:10.1128/AAC.01528-19. PubMed PMID: 31591124; PubMed Central PMCID: PMCPMC6879257.
- Sun C, Cui M, Zhang S, et al. Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008-2018. Emerg Microbes Infect. 2019;8(1):1524–1527. doi:10.1080/22221751.2019.1678367. PubMed PMID: 31631781; PubMed Central PMCID: PMCPMC6818123.
- He D, Wang L, Zhao S, et al. A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J Antimicrob Chemother. 2020 Feb 4. doi:10.1093/jac/dkaa012. PubMed PMID: 32016288.
- Wang L, Liu D, Lv Y, et al. Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, Eravacycline, and Omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob Agents Chemother. 2019 Dec 20;64(1). doi:10.1128/AAC.01326-19. PubMed PMID: 31611352.
- Lv L, Wan M, Wang C, et al. Emergence of a plasmid-Encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. mBio. 2020;11(2). doi:10.1128/mBio.02930-19.
- Wang X, Chen H, Zhang Y, et al. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: role of the global regulator RamA and its local repressor RamR. Int J Antimicrob Agents. 2015 Jun;45(6):635–640. doi:10.1016/j.ijantimicag.2014.12.022. PubMed PMID: 25681067. doi: 10.1016/j.ijantimicag.2014.12.022
- Poole K, Krebes K, McNally C, et al. Multiple antibiotic resistance in pseudomonas aeruginosa: evidence for involvement of an efflux Operon. J Bacteriol. 1993;175(22):7363–7372. doi: 10.1128/JB.175.22.7363-7372.1993
- Hou J, Wan W, Mao D, et al. Occurrence and distribution of sulfonamides, tetracyclines, quinolones, macrolides, and nitrofurans in livestock manure and amended soils of Northern China. Environ Sci Pollut Res Int. 2015 Mar;22(6):4545–4554. doi:10.1007/s11356-014-3632-y. PubMed PMID: 25318415. doi: 10.1007/s11356-014-3632-y
- Mohamed ER, Ali MY, Waly N, et al. The Inc FII plasmid and its contribution in the transmission of blaNDM-1 and blaKPC-2 in Klebsiella pneumoniae in Egypt. Antibiotics (Basel). 2019 Dec 13;8(4). doi:10.3390/antibiotics8040266. PubMed PMID: 31847288.
- Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018 Jul 4;7(1):122. doi:10.1038/s41426-018-0124-z. PubMed PMID: 29970891; PubMed Central PMCID: PMCPMC6030107.
- Ma K, Feng Y, Liu L, et al. A cluster of colistin- and carbapenem-resistant Klebsiella pneumoniae carrying blaNDM-1 and mcr-8.2. J Infect Dis. 2019 Dec 11. doi:10.1093/infdis/jiz519. PubMed PMID: 31822905.
- Hadjadj L, Baron SA, Olaitan AO, et al. Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front Microbiol. 2019;10:2720. doi:10.3389/fmicb.2019.02720. PubMed PMID: 31849875; PubMed Central PMCID: PMCPMC6887894. doi: 10.3389/fmicb.2019.02720
- Walsh TR, Wu Y. China bans colistin as a feed additive for animals. Lancet Infect Dis. 2016;16(10):1102–1103. doi:10.1016/s1473-3099(16)30329-2.