ABSTRACT
Nowadays, drug-resistant tuberculosis (DR-TB) and co-infected tuberculosis (CI-TB) strains are the leading cause for the enhancement of long-term morbidity and unpredicted mortality rates from this ghoulish acid fast-bacterium infection, globally. Unfortunately, the lack of/ample lethargic towards the development of compelling anti-TB regimens with a large-scale prevalence rate is a great challenge towards control of the pandemic situation. Indeed, the recent improvement in genomic studies for early diagnosis and understanding the mechanisms of drug resistance, as well as the identification of newer drug targets is quite remarkable and promising. Mainly, identification of such genetic factors, chromosomal mutations and associated pathways gives new ray of hope in current anti-TB drug discovery. This focused review provides molecular insights into the updated drug resistance mechanisms with encoded bacilli genetic factors as a novel target and potential source of development with screened-out newer anti-TB agents towards the control of MDR-TB soon.
Introduction
Mycobacterium tuberculosis (Mtb) is a grievous pathogenic organism, which has infected about one-third of the world’s population globally [Citation1,Citation2]. According to the World Health Organization (WHO) statistics, each year there were ∼ 8–10 million new tuberculosis (TB) cases continuously recorded [Citation1,Citation2]. African and Asian countries are suffering more than the Western Pacific region, so overall, 7–8 million new Mtb cases have emerged from African and Asian countries [Citation1]. In 2015, 60% of new Mtb cases had been recorded from India, Indonesia, China, Nigeria, Pakistan, and South Africa [Citation1]. The failure of first-line drugs (isoniazid or INH, rifampicin or RIF, ethambutol or EMB, pyrazinamide or PZA and streptomycin or STR), second-line drugs (moxifloxacin or MXF, kanamycin or KAN, capreomycin or CAP, ethionamide or ETH, para-aminosalicylic acid or PAS and cycloserine or DCS) and ineffectiveness of Directly Observed Treatment Short (DOTS) are the leading cause for the emergence of multi-drug resistant (MDR), extensively drug-resistant (XDR), extremely drug-resistant (XXDR) and ghoulish totally drug-resistant (TDR) strains, worldwide [Citation3]. Apart from that the increased rate of TB-HIV (Human Immunodeficiency Virus) co-infection is another issue leading to the pathetic situation towards the control of MDR-TB strain. In 2005, WHO reported ∼ 0.4 million death by TB-HIV co-infections [Citation1,Citation2].
In 1944, after the discovery of STR, TB treatment had been possible which created a revolution in anti-TB drug development. After that, there were several new classes of drugs introduced for treatment and recorded valid for a specific time against TB [Citation3,Citation4]. But nowadays, all mono and combination therapies, including DOTS and DOTS-Plus, are ineffective against the MDR-TB strains or bad bugs. Thus, TB still remains as one of the leading infectious diseases worldwide. Every year, it has reported 100,000 or 3.9% new cases of RIF-resistant TB from total 480,000 MDR-TB cases. Nowadays, data of XDR-TB cases from 117 countries suggested that around 9.5% MDR-TB changed to XDR TB [Citation2]. Approximately, 50%–70% TB cases are curable by current combinatorial chemotherapy, but it is not enough due to the spontaneous addition of new MDR-TB cases gradually. In comparison with other bacterial diseases, anti-TB drug resistance occurs just after the newly introduced chemotherapeutic agent. On the other hand, MDR-TB treatment rate is also expensive and time-consuming [Citation3,Citation4]. Currently used anti-TB drugs are not potential for killing the dormant and intracellular forms of Mtb. Thus, the present drug resistance situation gives a direct indication of searching the alternate and active anti-TB agent.
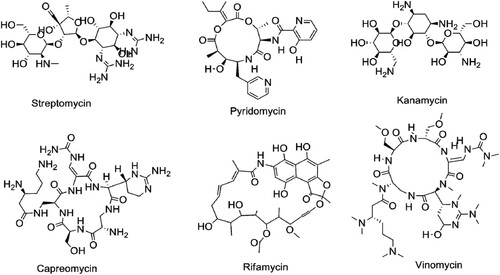
Since the last two decades, there has been a lot of progress to extract and identify the potent antimycobacterial chemicals/compounds from natural sources [Citation5,Citation6]. There are a lot of drugs discovered from the natural source such as STR and RIF []. Overall, natural sources are the main parental sources for the development of any semi-synthetic drug (RIF is a semi-synthetic-modified drug from rifamycin) [Citation7,Citation8]. WHO encourages and facilitates in national, regional and global level towards searching for new therapeutic agents for the eradication of TB in 2030. However, from the current status of anti-TB drug development record, WHO has changed their slogan “Stop TB to End TB.” With advancement in drug development techniques and instrumentation, there are several potential drug candidates developed with their novel mechanism of action including time-saving assays and ideal chemical isolation procedures compared to the previous drug discovery methods [Citation6–9]. Therefore, in this timeline review, we have discussed updated information on drug-resistant mechanisms of Mtb.
Drug-resistant mechanisms of Mtb
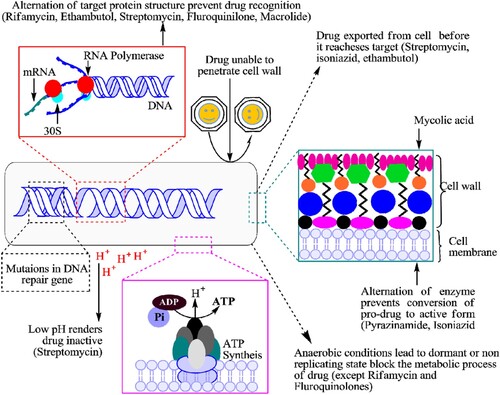
Currently, drug-resistant Mtb strains spread as per Darwin evolution theory by overcoming the anti-TB regimens through adopting genetic mutations and other mechanisms along with selective environmental pressure []. Both long-term unremitting combination drug therapies and environmental conditions have created an evolution of drug-resistant Mtb strains which become resistant gradually against the existing anti-TB drugs [Citation3,Citation9,Citation10]. Sometimes several environmental factors cause genetic modifications or mutations in the Mtb genome that not only reduce the effectiveness of the applied drug, but also provide the potential fitness for Mtb survival in any extreme condition [Citation10]. This survival represents the fatal exposures to bactericidal antibiotics through radical-induced mutagenesis and promotes the multidrug-resistant phenotypes of Mtb. Sometimes, if this reactive oxygen and free-radicals fail to eradicate the mycobacterial cell, then bad promotion of cell mutagenesis and the rise of drug resistance [Citation3,Citation10].
Mostly, TB infection occurs through the engulfment of the bacterium by dendrite cells or alveolar macrophages, where bacilli avoid the killing mechanism and remain to reproduce evading phagosome-lysosome membrane fusion [Citation11,Citation12]. Complementary macrophages and other immune cells are usually contained in a specific site, known as granuloma. Particularly in the granuloma, Mtb bacilli are replicating vigorously along with non-replicating persistent/dormant form of Mtb in the specific site, which is prompted by the environmental circumstances, associated with anorexia/ hypoxia, nitric oxide production, and nutrient deprivation [Citation11,Citation13].
Intrinsic resistance mechanisms in Mtb
The intrinsic mechanism of Mtb is resistance not only to the existing drugs but also to newly introduced drugs []. Broadly, the drug-resistant mechanisms of Mtb are divided into two categories, such as intrinsic and acquired for passively neutralizing the activity of applied anti-TB drugs/regimens [Citation3,Citation10]. Mtb attributed natural resistance mechanism against the macrolide class of antibiotics to low cell wall permeability through the expression of emr37. Particularly, emr37 belongs to such type of genetic factors that organize 23S rRNA-binding site through methylation [Citation10,Citation13]. Similarly, for natural resistance mechanism, the minimum inhibitory concentration (MIC) value of clarithromycin had decreased eight- to four-fold, by a permeability signalling barrier in the Mtb cell [Citation10]. Using the intrinsic drug resistance, Mtb has continually developed the unfamiliar structure of cell wall that is related to low permeability against applying anti-TB chemotherapy [Citation14].
In a study, the Mtb-Rv1698 has been directly associated with intrinsic resistance to hydrophilic antimycobacterial activities due to MspA genetic factors which is the porin-associated factor present in the Mtb cell wall that leads to lower the drug permeability [Citation11,Citation14]. The auto-regulator transcriptional activator WhiB7 controls the intrinsic drug resistance through the multidrug transporter tap, a transporter responsible for mycobacterial efflux against tetracycline or TET, STR and PAS, after binding with the somatic sigma factor, SigA [Citation12,Citation13]. Mainly, WhiB7, a small protein structure, contains 122 amino acids and an iron-sulphur group, which not only regulates the antibiotic resistance gene-like, eis, erm37 and tap, but also controls oxi-reductive reagents like dithiothreitol and diamide [Citation12]. Subsequently, the eukaryotic-like protein kinase G (PknG), a type of virulence factor which is involved in the survival of Mtb in host-macrophages by regulating the redox-homeostatic system. Similarly, the nucleoid-associated transcription factor Lsr2 controls the oxygen level for the persistence of Mtb in host-cell through the expression of iniBAC, a promoter plays a crucial role during the inhibition of cell wall biosynthesis and EfpA, a transport protein associated with efflux mechanisms [Citation14]. Additionally, the chromosomal protein Mar-regulon or MarA is also reducing the susceptibility of antibiotics like TET through regulating the MDR efflux pump. The overall antibiotic sensitivity and cellular redox status depend on the expression of NADH and reflux pump-associated transcription factors [Citation12–15].
Thus, stress-responsive sigma factors (SigA, SigF), associated transcriptional proteins (MarA, SoxR and Rob) along with several specific oxidative stress, are significantly associated with the resistance of applied anti-TB drugs [Citation3,Citation11,Citation16]. Several bioinformatic approaches have revealed the relationship between Mtb outer membrane proteins with intrinsic resistance against antibiotics in proteomic level [Citation11,Citation14]. However, apart from the permeability barriers of Mtb, physiological adaptations within the host can also manage antibiotic resistance [Citation11,Citation17]. Thus, understanding the intrinsic resistance is one of the major aspects in the Mtb drug development; because few drugs are available for treatment and could be a control in an iron hand of those gruesome Mtb strains by targeting intrinsic resistance-associated genetic factors [Citation18,Citation19].
Acquired resistance mechanisms in Mtb
Acquired drug resistance is commonly facilitated by the horizontal transfer of mobile genetic elements such as plasmids and transposons. Acquired drug resistance occurs in two different ways, such as chromosomal or gene mutation mechanisms and extra-chromosomal or gene transfer mechanisms [Citation3,Citation10,Citation19]. However, in Mtb, no such type of horizontal transfer of drug resistance genes has been reported, but most of them were found to be due to the origin of chromosomal mutations under selective pressure. Microorganisms also pay a physiological cost for drug resistance against anti-TB regimens. As a result, the rate of mutations in base pair is inversely genome size closely 0.0033 per replication in prokaryotes; however, in Mtb cases, most of the lead anti-TB regimens occur at 10−9 mutations per cell division [Citation16,Citation20]. Thus, the nature of drug selection is also directly related to the rate of mutation and that is the main ideal approach behind the combined formulation of anti-TB regimen in every single dose. The fitness cost of individual microorganisms depends on their growth and virulence transmission capacity from one host to another. For example, mutations on rpoB gene manage RIF-resistance in Mtb clinical isolates []; but sometimes the same isolates have less fitness cost in vitro. Thus, it may depend on mutations associated with some minor or major cost of the fitness of a strain [Citation10]. Therefore, fitness cost depends on the specific resistance mutation and the genetic circumstances of the strain.
Figure 3. A schematic representation of genetic mutations associated with the drug-resistant mechanism of rifampicin targeting rpoB.
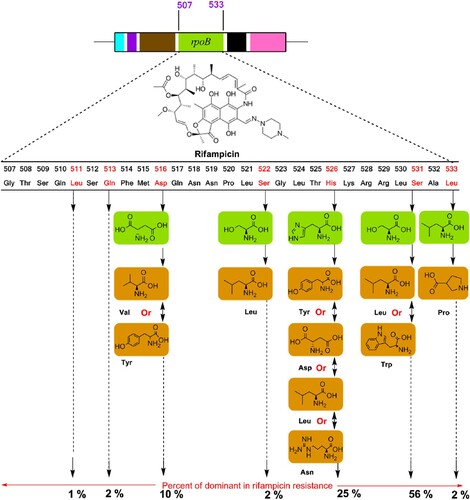
The fitness cost of various chromosomal mutations is directly proportional to antibiotic resistance, but there are limited data available on the resistance of drug and the fitness cost in Mtb. However, a chromosomal mutation is the most well-known mechanism of an antibiotic resistance [Citation3,Citation16,Citation19]. Mutation in S315T on the catalase-peroxidase enzyme (katG) is the most pervasive mutation and nearly 40%–94% of resistance is associated with INH-resistant Mtb clinical isolates [Citation3,Citation10]. Mutations are capable of reducing the ability of katG to convert INH into iso-nicotinic acid, a precursor for the formation of the INH-NAD adduct in mycolic acid synthesis pathways [Citation3,Citation10]. However, RIF-resistant clinical isolates have been compared with their susceptible parental strain, four out of five strains with the mutation S531L and no fitness cost for this mutation. Similarly, mutations in the promoter region of enoylacyl carrier protein reductase (inhA) are associated with the over-expression of inhA which is the main cause of INH resistance. The frequently detected mutation at the position of C15T in inhA regulatory region is associated with INH resistance with MIC <1 μg/mL [Citation21]. Only one plasmid is enough for carrying the resistance-encoded gene to overcome several drugs, the resistance of chloramphenicol, STR and TET are a suitable example [Citation22]. Overall, antibiotics become ineffective in 10%–20% of the cases through chromosomal changes and ∼ 80% of cases of resistance by the extra-chromosomal process [Citation22].
Drug-resistant mechanisms of anti-TB drugs
Several newly introduced drugs were reportedly ineffective due to the emergence of MDR-TB, XDR-TB strains [Citation3,Citation4,Citation23]. The current anti-TB regimens managed Mtb strains but not on a long-term basis as compared to STR. Cumulatively, environmental conditions, antibiotic pressure and chromosomal mutations play a major role in the development of drug resistant Mtb strain and transmission of those strains into a new host [Citation10,Citation13,Citation16].
Resistance mechanism of drug targeting cell wall, mycolic acid and folic acid synthesis
The Mtb cell wall comprises three covalently attached macromolecules, such as peptidoglycan, arabinogalactan and mycolic acid, known as “mycolyl-arabinogalactan-peptidoglycan” or mAGP complex [Citation13,Citation18,Citation19]. The role of mycolic acid was well known and several anti-tubercular drugs were developed by targeting it and its associated enzyme in the biosynthesis, such as INH and EMB, used as a prodrug towards inhibition/alternation of mycolic acid biosynthesis targeting encoded enzymes, katG and inhA [Citation10,Citation22,Citation24]. Particularly, INH makes a covalent adduct with the nicotinamide adenine dinucleotide (NAD) cofactor and that adduct complex acts as a tight-binding competitive inhibitor of inhA. However, mutations in the enzyme katG and inhA confer resistance to both EMB and INH [Citation28]. S315T mutation of katG was considered as the universal mutation and nearly 40–94% INH resistance recorded in newer Mtb isolates [Citation20]. Using advanced molecular tools, several other associated genetic factors related to cell wall biosynthesis (kasA, AhpC, niA, FadE24, ndh and FabG1) were explored in relation to anti-TB drug resistance [Citation25,Citation26]. Similarly, mutations in embB and embC cause resistance by Robinson transfers that are also associated with mycobacterial cell wall synthesis [Citation27]. Recently, mutations in embB along with mutations in decaprenyl-phosphoryl-5-phosphoribose (DPPR) synthase gene (ubiA), are associated with a high level of EMB resistance. The cell wall targeting anti-TB drug, DCS, clofazimine, amoxicillin, meropenem, imipenem and thioacetazone have been recorded ineffective due to the resistance development by mutation with individual genetic factors, depicted in [Citation28–53]. PAS is one of the most effective anti-TB drugs against multidrug-resistant tuberculosis since 1994 by targeting folic acid biosynthesis-associated enzyme, dihydropteroate synthase (DHPS) and iron metabolism inhibition. Nowadays, PAS has been ineffective due to mutation in the genetic factor, thyA gene (encoding with thymidylate synthase) and folC gene (dihydrofolate synthase or DHFS) [Citation54]. Particularly, a mutation at Thr202Ala in thyA gene is most common for ∼ 37% of PAS-resistant strains [Citation54,Citation55]. Newly introduced two drugs, delamanid and pretomanid, have been more effective towards the inhibition of Mtb cell wall synthesis in the last couple of years; but recently, drug resistance against these two drugs have been reported across the glob [Citation54–56].
Table 1. Genetic factors involved in first-line anti-TB drug resistance in Mycobacterium tuberculosis.
Table 2. Genetic factors involved in second-line anti-TB drug resistance in Mycobacterium tuberculosis.
Table 3. Genetic factors involved in third-line anti-TB drug resistance in Mycobacterium tuberculosis.
Several chemicals have been designed to target the aminoacyl-tRNA synthetases (AARS)-associated enzymes which are essential enzymes required for bacterial protein synthesis [Citation57]. Moreover, some unique lipids are present in the Mtb cell envelope and act as a permeability barrier to use anti-TB drugs. As a result the lipid barrier plays an essential role in Mtb drug resistance [Citation58]. The structurally distinct mycobacterial lipids, derived from malonyl coenzyme A (CoA) are assembled with acyl-CoA carboxylases (ACC) subunits, AccA1 to AccA3, AccD1 to AccD6, AccE5. Moreover, ACC is an essential factor for fatty acids, mycolates and lipid synthesis of Mtb along with biotin (vitamin H or vitamin B7), as the cofactor in the post-translational process [Citation59–62]. Thus, de novo biotin biosynthesis or biotin-ACC ligation inhibition is a new target for Mtb inhibition.
Resistance mechanism of drug targeting ATP, DNA, RNA and protein synthesis
The pandemic Mtb-resistant traits arising through evolution, antibiotic pressure and other means which deliver this mutant feature for their survival in a new environment [Citation20,Citation21,Citation36]. Due to a point mutation, the applied drugs cannot bind the proper sites of a target and as a result decrease the potency [Citation10,Citation13,Citation14,Citation17]. The central dogma pathways, such as DNA, RNA and protein synthesis, are encoded nodal enzymes and precursors to achieve the required integration for surviving in an environment or a host body. Thus, several anti-TB drugs are developed targeting ATP, DNA and protein synthesis, because these are crucial to produce energy for the survival of Mtb [Citation3,Citation10,Citation16]. Diarylquinoline and imidazopyridine amide class of antibiotics/drugs are targeting for the inhibition of ATP synthesis of Mtb, PAS inhibits DNA precursor, and fluoroquinolone class of antibiotics inhibits DNA gyrase of Mtb [Citation48]. RIF inhibits RNA synthesis, while oxazolidinones (linezolid), aminoglycosides, macrolides, and cyclic peptides inhibit protein synthesis, among them linezolid is the potential orally existing drug targeting Mtb protein synthesis. Mutated rpoB encourages a conformational change towards the binding affinity of RIF at β-subunit of the RNA polymerase, and the drug became inactive without proper binding to the exact target site [Citation30,Citation31; ].
Future prospective of drug resistance and drug development
For a long time, there is a duel between humans and bacteria for surviving from each other. The development of antibiotics changed the whole thing for several decades. Nowadays, Mtb produce more unique genetic factors and develop quorum sensing bio-film mechanism to counter-attack against the new anti-TB drug [Citation14–17]. Modern mutants can regain fitness for enhancing transmissibility of virulence from adopting a resistance mechanism from the existing mutant strains with a suitable environmental pressure [Citation20,Citation23,Citation24]. In the past, several MDR epidemic analyses confirmed that evolution of regulatory systems by an antibiotic during the transition of Mtb from a dormant state to active growth and increased the phenotypic drug tolerance of latent TB.
In the current perspective, drug-resistant TB is associated with genetic mutations on the Mtb target enzyme [Citation27,Citation34,Citation44,Citation47,Citation55]. The resistance factor related to variations is broadly divided into two types i.e. cellular mechanisms connected with mismatch renovation, DNA polymorphisms error, translational inaccuracy, etc., and external mechanisms connected with stress factor, host-environment, pollution, quality of diagnosis, etc. [Citation17–19]. Additionally, inadequate hospital facility, inappropriate guidance, long-term hospital expenses, lack of strict national antibiotic policy, awareness, illiteracy and poverty are some of the associated social-economic factors in drug-resistant TB [Citation63,Citation64]. In short, the non-cooperation of TB patients and costs are directly affecting to achieve the complete cure. Thus, TB is one of the primary causes of death in the Asian population and the more common cause of adult mortality than HIV, malaria and other tropical infectious diseases [Citation1,Citation4,Citation65]. The development of co-infection of TB with HIV has lead to the increase in MDR-TB, MDR-TB and XDR-TB strains [Citation1,Citation2]. Mostly, rates of TB incidence tripled in African countries with high HIV prevalence. Approximately 13% of HIV-TB cases were recorded from global TB cases; among them 39% HIV-TB from Africa [Citation1,Citation2]. As a result, complex resistance profiles with intersecting toxicity and a substantial amount of drug consumption obstruct towards the control of co-infected TB.
Continuously approved anti-TB drugs bedaquiline, delamanid and pretomanid in a small duration encouraging the drug development performance. But we need to develop more and more to control millions of TB-infected people, globally [Citation4,Citation8,Citation48–50]. Approximately, ∼ 265 billion dollars for the control of TB as for the development of a new diagnosis and treatment in the medical research, globally, but the output is only 0.25% [Citation4,Citation8,Citation66]. From WHO report, there are several anti-TB agents in the pipeline and hope for a discovery in the recent future. According to the current reports, most tubercular agents contain nitrogen heterocyclic and pyridine and pyrazine derivatives similar to the existing anti-TB drug, INH, PZA, etc. Precisely, nitrogen heterocyclic containing nucleus is an essential chemical scaffold for a future anti-TB drug with changed physicochemical, metabolic and pharmacokinetic properties, through the addition of a potential side chain molecule [Citation67,Citation68].
The excellent progress screening procedure with possible adopted techniques in medicine and molecular mechanisms is expected to the development of the effective anti-TB drug in recent future []. Besides, towards searching for a new anti-TB regimen, natural phyto-metabolites (secondary plant metabolites) and phyco-chemical (cyanobacterial compound) have high attention due to their potential reported towards promoting health and reducing disease burden [Citation5–8]: terpenoid class of phytochemicals, mono-O-methyl curcumin isoxazole from well-known Indian medicinal plant, Curcuma longa with the MIC value, 00019 mg/mL and abietane from a Chinese sub-species, Plectranthus grandidentatus, with the MIC value 0.00039 mg/mL, the quinone class of phyto-compound, plumbagin from Diospyros anisandra, with the MIC value, 0.0015–0.0033 mg/mL and 7-methyljuglone from the South African medicinal shrub Euclea natalensis with the MIC value 00057 mg/mL, an organic heterocyclic compound, calanolide A from the Malaysian sub-species, Calophyllum lanigerum, with the MIC value, 0.0031–0.016 mg/mL, the alkaloid class of constitution, dihydro-β-agarofuran sesquiterpene from Celastrus vulcanicola, with 0.0062 mg/mL [Citation69–72].
Similarly, several unique bioactive marine alkaloid class of products, such as Ecteinascidin 770 from the sea squirt, Ecteinascidia thurstoni with the MIC value 0.00013 mg/mL, Hirustellone A-D from the fungi, Hirustella nivea BCC2594 with the MIC value 0.00078 mg/mL, 8-Hydroxymanzamine J from marine sponge Acanthostrongylophora sp., with the MIC value 0.00040 mg/mL were recorded against Mtb [Citation73,Citation74]. Additionally, the phenolic class of chemical nocardithiocin from marine bacterium Nocardia pseudo brasiliensis with the MIC value 0.00002–0.00115 mg/mL, Trichoderin A from marine fungi Trichoderma sp., with the MIC value, 0.00012 mg/mL, Lariatin A from marine bacterium Rhodococcus jostii with the MIC value 0.00039 mg/mL were recorded as most preferable anti-TB candidates [Citation75–77].
In comparison, plant chemicals are ideal drug candidates than the marine chemicals in the host-toxicity point of view; however, the drug success rate is higher in marine products than plant products [Citation5,Citation6]. Thus, an ideal drug candidate should manage both activity and toxicity with pharmacokinetic properties for more success in drug validation modules. Strategically, several advanced chemical conjugation/drug modification through medicinal chemistry protocol and nanoparticle/nanocarrier formulations has been adopted for the utilization of more natural products in anti-TB drug development [Citation78–80]. Nevertheless, long-term investment, administrative commitment, interior geographical support and scientific endeavour will be playing the crucial factors for a sustainable TB-free society.
Conclusion
The prevalence rate of Mtb and resistance mechanisms against ongoing therapy is a serious global concern. The absence of potential drug candidates and patient awareness along with unhygienic practices are the prime reasons for rapid emergence of MDR, TDR, XDR Mtb strains. Understanding and analysis of compensatory evolution, clonal interference, declined cell wall permeability, overexpression of efflux pumps, target modification and target mimicry modulates are the key factors associated with drug resistance. The powerful molecular mechanism with complex pathway information of drug resistance of Mtb immensely requires counter-attacking towards the development of new anti-TB regimens. Hopefully, the combination of new genomics knowledge of drug-resistant mechanisms in Mtb would give a new direction for combination drug discovery and tremendous support for the production of highly effective anti-TB drugs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- World Health Organization. Global tuberculosis report-2018.
- World Health Organization. MDR TB fact sheet-2016.
- Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis. 2015;19:1276–1289.
- Koul A, Arnoult E, Lounis N, et al. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490.
- Santhosh RS, Suriyanarayanan B. Plants: a source for new antimycobacterial drugs. Planta Med. 2014;80:9–21.
- Liu X, Chen C, He W, et al. Exploring anti-TB leads from natural products library originated from marine microbes and medicinal plants. Antonie Van Leeuwenhoek. 2012;102:447–461.
- Swain SS, Paidesetty SK, Padhy RN, et al. Computational approach for locating effective cyanobacterial compounds against Mycobacterium tuberculosis. Indian J Pharmac Edu Res. 2017;51:1–10.
- Sloan DJ, Davies GR, Khoo SH. Recent advances in tuberculosis: new drugs and treatment regimens. Curr Respir Med Rev. 2013;9:200–210.
- Mitchison DA, Davies GR. Assessment of the efficacy of new anti-tuberculosis drugs. Open Infect Dis J. 2008;2:59–76.
- Palomino JC, Martin A. Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics (Basel). 2014;3:317–340.
- Siroy A, Mailaender C, Harder D, et al. Rv1698 of Mycobacterium tuberculosis represents a new class of channel-forming outer membrane proteins. J Biol Chem. 2008;283:17827–17837.
- Burian J, Ramón-García S, Howes CG, et al. Whib7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev Anti Infect Ther. 2012;10:1037–1047.
- Kashyap A, Singh PK, Silakari O. Mechanistic investigation of resistance via drug-inactivating enzymes in Mycobacterium tuberculosis. Drug Metab Rev. 2018;50:448–465.
- Viveirosa M, Martins M, Rodrigues L, et al. Inhibitors of mycobacterial efflux pumps as potential boosters for anti-tubercular drugs. Expert Rev Anti Infect Ther. 2012;10:983–998.
- Bartek IL, Woolhiser LK, Baughn AD, et al. Mycobacterium tuberculosis Lsr2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. mBio. 2014;5:e01106–14. doi:10.1128/mBio.01106-14.
- Morris RP, Nguyen L, Gatfield J, et al. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2005;102:12200–12205.
- Nguyen L, Pieters J. Mycobacterial subversion of chemotherapeutic reagents and host defense tactics: challenges in tuberculosis drug development. Annu Rev Pharmacol Toxicol. 2008;49:427–453.
- Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic-a vision for applied use. Biochem Pharmacol. 2006;71:910–918.
- Smith T, Wolffv KA, Nguyen L. Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr Top Microbiol Immunol. 2013;374:53–80.
- Lynch M. The lower bound to the evolution of mutation rates. Genome Biol Evol. 2011;3:1107–1118.
- Aye KS, Nakajima C, Yamaguchi T, et al. Genotypic characterization of multi-drug-resistant Mycobacterium tuberculosis isolates in Myanmar. J Infect Chemother. 2016;22:174–179.
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260.
- Gygli SM, Borrell S, Trauner A, et al. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. 2017;41:354–373.
- Almeida Da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother. 2011; 66: 1417-1430.
- Ando H, Kitao T, Miyoshi-Akiyama T, et al. Down regulation of katG expression is associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 2011;79:1615–1628.
- Cardoso RF, Cardoso MA, Leite CQ, et al. Characterization of ndh gene of isoniazid resistant and susceptible Mycobacterium tuberculosis isolates from Brazil. Mem Inst Oswaldo Cruz. 2007;102:59–61.
- Xu Y, Jia H, Huang H, et al. Mutations found in embCAB, embR, and ubiA genes of ethambutol-sensitive and -resistant Mycobacterium tuberculosis clinical isolates from China. Bio Med Res Int. 2015;951706; doi:10.1155/2015/951706.
- Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol. 2006;62:1220–1227.
- Pourakbari B, Mamishi S, Mohammadzadeh M, et al. First-line anti-tubercular drug resistance of Mycobacterium tuberculosis in Iran: a systematic review. Front Microbiol. 2016;28:1139, doi:10.3389/fmicb.2016.01139.
- Sensi P. History of the development of Rifampin. Rev Infect Dis. 1983;5:S402–S406.
- Campbell EA, Korzheva N, Mustaev A, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912.
- Forbes M, Kuck NA, Peets EA. Mode of action of ethambutol. J Bacteriol. 1962;84:1099–1103.
- Konno K, Feldmann FM, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95:461–469.
- Zhang S, Che J, Shi W, et al. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect. 2013;2:e34, doi:10.1038/emi.2013.38.
- Honoré N, Cole ST. Streptomycin resistance in mycobacteria. Antimicrob Agent Chemother. 1994;38:238–242.
- Dookie N, Rambaran S, Padayatchi N, et al. Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. J Antimicrob Chemother. 2018;73:1138–1151.
- Rustomjee R, Lienhardt C, Kanyok T, et al. A phase II study of the sterilising activities of ofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int J Tuberc Lung Dis. 2008;12:128–138.
- Yanagisawa K, Sato N. Studies on kanamycin, a new antibiotic against tubercle bacilli. I. Effect on virulent tubercle bacilli in vitro and in mice. J Antibiot (Tokyo). 1957;10:233–235.
- Stark WM, Higgens CE, Wolfe RN, et al. Capreomycin-a new antimycobacterial agent produced by Steptomyces capreolus sp. Antimicobacter Agent Chemother. 1963;1962:596–606.
- Lin SY, Rodwell TC, Victor TC, et al. Pyrosequencing for rapid detection of extensively drug-resistant Mycobacterium tuberculosis in clinical isolates and clinical specimens. J Clin Microbiol. 2014;52:475–482.
- Thee S, Garcia-Prats AJ, Donald PR, et al. A review of the use of ethionamide and prothionamide in childhood tuberculosis. Tuberculosis (Edinb). 2016;97:126–136.
- Lehmann J. Para-aminosalicylic acid in the treatment of tuberculosis. Lancet. 1946;1:15.
- Cáceres NE, Harris NB, Wellehan JF, et al. Overexpression of the D-alanine racemase gene confers resistance to D-cycloserine in Mycobacterium smegmatis. J Bacteriol. 1997;179:5046–5055.
- Chen JM, Uplekar S, Gordon SV, et al. A point mutation in cycA partially contributes to the D-cycloserine resistance trait of Mycobacterium bovis BCG vaccine strains. PLoS One. 2012;7:e43467, doi:10.1371/journal.pone.0043467.
- Yano T, Kassovska-Bratinova S, Teh JS, et al. Reduction of clofazimine by mycobacterial type 2 NADH: quinone oxidoreductase: a pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem. 2011;286:10276–10287.
- Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through up regulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agent Chemother. 2014;58:2979–2981.
- Almeida D, Ioerger T, Tyagi S, et al. Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob Agent Chemother. 2016;60:4590–4599.
- Palomino JC, Martin A. TMC207 becomes bedaquiline, a new anti-TB drug. Fut Microb. 2013;8:1071–1080.
- Upton AM, Cho S, Yang TJ, et al. In vitro and in vivo activities of the nitroimidazole TBA-354 against Mycobacterium tuberculosis. Antimicrob Agent. Chemother. 2015;59:136–1344.
- Manjunatha UH, Boshoff H, Dowd CS, et al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:431–436.
- Feuerriegel S, Koser CU, Bau D, et al. Impact of Fgd1 and ddn diversity in Mycobacterium tuberculosis complex on in vitro susceptibility to PA-824. Antimicrob Agents Chemother. 2011;55:5718–5722.
- Bozdogan B, Appelbaum PC. Oxazolidinones: activity, mode of action, and mechanism of resistance. Int J Antimicrob Agent. 2004;23:113–119.
- Richter E, Rüsch-Gerdes S, Hillemann D. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agent Chemother. 2007;51:1534–1536.
- Zhao F, Wang XD, Erber LN, et al. Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob Agent Chemother. 2014;58:1479–1487.
- Mathys V, Wintjens R, Lefevre P, et al. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2009;53:2100–2109.
- Sharma D, Sharma S, Sharma J. Potential strategies for the management of drug resistant tuberculosis. J Glob Antimicrob Resist. 2020; doi:10.1016/j.jgar.2020.02.029.
- Palencia A, Li X, Bu W, et al. Discovery of novel oral protein synthesis inhibitors of Mycobacterium tuberculosis that target leucyl-trnasynthetase. Antimicrob Agent Chemother. 2016;60:271–280.
- Nguyen L, Pieters J. Mycobacterial subversion of chemotherapeutic reagents and host defence tactics: challenges in tuberculosis drug development. Annu Rev Pharmacol Toxicol. 2009;49:427–453.
- Tiwari D, Park SW, Essawy MM, et al. Targeting protein biotinylation enhances tuberculosis chemotherapy. Sci Transl Med. 2018;10:pii: eaal1803, doi:10.1126/scitranslmed.aal1803.
- Lyonnet BB, Diacovich L, Cabruja M, et al. Pleiotropic effect of AccD5 and AccE5 depletion in acyl-coenzyme A carboxylase activity and in lipid biosynthesis in mycobacteria. PLOS One. 2014;9:e99853, doi:10.1371/journal.pone.0099853.
- Oh TJ, Daniel J, Kim HJ, et al. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA carboxylase in Mycobacterium tuberculosis H37Rv. J Biol Chem; 2006;281:3899–3908.
- Cholo MC, Mothiba MT, Fourie B, et al. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J Antimicrob Chemother. 2017;72:338–353.
- Bhatt R, Chopra K, Vashisht R. Impact of integrated psycho-socio-economic support on treatment outcome in drug resistant tuberculosis-a retrospective cohort study. Indian J Tuberc. 2019;66:105–110.
- Muniyandi M, Rao VG, Bhat J, et al. Health literacy on tuberculosis amongst vulnerable segment of population: special reference to Saharia tribe in central India. Indian J Med Res. 2015;141:640–647.
- Sizemore CF, Schleif AC, Bernstein JB, et al. The role of biomedical research in global tuberculosis control: gaps and challenges: a perspective from the US National Institute of Allergy and Infectious Diseases, National Institutes of Health. Emerg Microbes Infect. 2012;1:e9, doi:10.1038/emi.2012.21.
- Beena RD. Antituberculosis drug research: a critical overview. Med Res Rev. 2013;33:693–764.
- Kumar K, Awasthi D, Lee SY, et al. Novel tri-substituted benzimidazoles, targeting Mtb FtsZ, as a new class of antitubercular agents. J Med Chem. 2011;54:374–381.
- Reddy VM, Reddy KR. Synthesis and antimicrobial activity of some novel 4-(1H-benz[d]imidazol-2yl)-1, 3-thiazol-2-amines. Chem Pharm Bull. 2010;58:953–956.
- Xu ZQ, Barrow WW, Suling WJ, et al. Anti-HIV natural product (+)-calanolide A is active against both drug-susceptible and drug-resistant strains of Mycobacterium tuberculosis. Bioorg Med Chem. 2004;12:1199–1207.
- Uc-Cachón AH, Borges-Argáez R, Said-Fernández S, et al. Naphthoquinones isolated from Diospyros anisandra exhibit potent activity against pan-resistant first-line drugs Mycobacterium tuberculosis strains. Pulm Pharmacol Ther. 2014;27:114–120.
- Lall N, Meyer JJM, Wang Y, et al. Characterization of intracellular activity of antitubercular constituents the roots of Euclea natalensis. Pharm Biol. 2005;43:353–357.
- Changtam C, Hongmanee P, Suksamrarn A. Isoxazole analogs of curcuminoids with highly potent multidrug-resistant antimycobacterial activity. Eur J Med Chem. 2010;45:4446–4457.
- Suwanborirux K, Charupant K, Amnuoypol S, et al. Ecteinascidins 770 and 786 from the Thai tunicate Ecteinascidia thurstoni. J Nat Prod. 2002;65:935–937.
- Isaka M, Prathumpai W, Wongsa P, et al. Hirsutellone F, a dimer of antitubercular alkaloids from the seed fungus Trichoderma sp. BCC 7579. Org Lett. 2006;8:2815–2817.
- Mukai A, Fukai T, Hoshino Y, et al. Nocardithiocin, a novel thiopeptide antibiotic, produced by pathogenic Nocardia pseudo brasiliensis IFM 0757. J Antibiot. 2009;62:613–619.
- Iwatsuki M, Tomoda H, Uchida R, et al. Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J Am Chem Soc. 2006;128:7486–7491.
- Pruksakorn P, Arai M, Kotoku N, et al. Trichoderins, novel aminolipopeptides from a marine sponge-derived Trichoderma sp., are active against dormant mycobacteria. Bioorg Med Chem Lett. 2010;20:3658–3663.
- Nasiruddin M, Neyaz MK, Das S. Nanotechnology based approach in tuberculosis treatment. Tuberc Res Treat. 2017: 1–12. doi:10.1155/2017/4920209.
- Swain SS, Paidesetty SK, Dehury B, et al. Computer-aided synthesis of dapsone-phytochemical conjugates against dapsone-resistant Mycobacterium leprae. Sci Rep. 2020a;10; doi:10.1038/s41598-020-63913-9.
- Swain SS, Paidesetty SK, Padhy RN, et al. Isoniazid-phytochemical conjugation: a new approach for potent and less-toxic anti-TB drug development. Chem Biol Drug Des. 2020b;1–17. doi:10.1111/cbdd.13685.