ABSTRACT
The global pandemic of coronavirus disease 2019 (COVID-19) is a disaster for human society. A convenient and reliable neutralization assay is very important for the development of vaccines and novel drugs. In this study, a G protein-deficient vesicular stomatitis virus (VSVdG) bearing a truncated spike protein (S with C-terminal 18 amino acid truncation) was compared to that bearing the full-length spike protein of SARS-CoV-2 and showed much higher efficiency. A neutralization assay was established based on VSV-SARS-CoV-2-Sdel18 pseudovirus and hACE2-overexpressing BHK21 cells (BHK21-hACE2 cells). The experimental results can be obtained by automatically counting the number of EGFP-positive cells at 12 h after infection, making the assay convenient and high-throughput. The serum neutralizing titer measured by the VSV-SARS-CoV-2-Sdel18 pseudovirus assay has a good correlation with that measured by the wild type SARS-CoV-2 assay. Seven neutralizing monoclonal antibodies targeting the receptor binding domain (RBD) of the SARS-CoV-2 S protein were obtained. This efficient and reliable pseudovirus assay model could facilitate the development of new drugs and vaccines.
Introduction
As of August 18th, 2020, over 21.9 million cases of coronavirus disease 2019 (COVID-19) have been reported. The spread of the SARS-CoV-2 is still difficult to control due to its highly contagious nature and asymptomatic infection. An urgent need exists to develop vaccines or therapeutics against SARS-CoV-2 infection. However, SARS-CoV-2 culture and assays need to be carried out in a biosafety level-3 laboratory, which limits the efficiency of research and development (R & D). Widely available and convenient functional assays, such as cell-based assays for neutralizing quantification, are essential for vaccine and therapeutic antibody R & D [Citation1–3].
SARS-CoV-2 belongs to the genus betacoronavirus and is an enveloped virus [Citation4]. The glycosylated spike (S) protein is the major surface protein and is responsible for receptor binding and the fusion of the viral membrane with cellular membranes [Citation5,Citation6]. Studies have shown that SARS-CoV-2 S protein and SARS-CoV S protein bind with the same receptor, human angiotensin-converting enzyme 2 (hACE2), with similar affinities [Citation7–9]. Therefore, studies of SARS-CoV neutralization assays, which are based on pseudoviruses bearing the spike protein of SARS-CoV, would provide valuable experience and reference for SARS-CoV-2 assays. In recent years, a pseudotyping system based on vesicular stomatitis virus (VSV) was reported to produce pseudotyped viruses incorporating the envelope protein of heterologous risk group-3 (RG-3) or RG-4 viruses, such as Ebola virus, SARS coronavirus, and MERS coronavirus, in which the VSV G gene is deleted (VSVdG) and the gene encoding GFP, luciferase or other reporter genes was integrated [Citation10–12]. Pseudotyped viruses provide a safe viral entry model because of their inability to produce infectious progeny virus. VSV assembly occurs at the plasma membrane and involves the budding of virions from the cell surface. During budding, VSV acquires an envelope, consisting of a lipid bilayer derived from the plasma membrane, and spike proteins, consisting of trimers of the VSV glycoprotein (VSV-G). When VSV-G is absent and the glycoprotein from the heterologous virus is complacently expressed in cells infected with VSVdG, the glycoprotein of the heterologous virus could be assembled into the VSV membrane.
Recently, Michael Letko et al. used VSVdG-luc bearing SARS-spike chimeras to study cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses [Citation13]. Jianhui Nie et al. successfully constructed a pseudovirus neutralization assay for SARS-CoV-2, which consists of pseudotyped VSV bearing the full-length spike protein of SARS-CoV-2 and Huh7 cells [Citation12]. A lentiviral pseudotype bearing the truncated spike protein of SARS-CoV-2 was also constructed and used to study virus entry and immune cross-reactivity with SARS-CoV. Viral packaging and infection efficiency are limiting factors for high-throughput in vitro neutralization assays.
It was reported that a pseudotyped VSV and a retrovirus bearing a SARS-CoV-S protein variant with a truncation in the cytoplasmic tail had much higher infection efficiency than that with full-length S protein [Citation11, Citation14]. In this study, VSVdG pseudotypes with full-length SARS-CoV-2 S protein or truncated SARS-CoV-2-Sdel18 protein with a C-terminal 18 aa truncation were compared, and we found that the infection efficiency of VSV-SARS-CoV-2-Sdel18 was much higher than that of VSV-SARS-CoV-2-S. Among the cell lines used in this study, BHK21-hACE2 cells stably expressing human ACE2 were most sensitive to infection, while Vero-E6 cells were most suitable for viral packaging. A neutralization assay for antibody screening and validation was established based on VSV-SARS-CoV-2-Sdel18 and BHK21-hACE2 cells, and 7 strains of neutralizing monoclonal antibodies targeting the receptor binding domain (RBD) of SARS-CoV-2 S protein were obtained. Most importantly, the neutralizing antibody titers measured by the pseudovirus assay and the authentic virus assay were highly correlated.
Materials and methods
Cells and samples
Vero-E6 (American Type Culture Collection [ATCC], CRL-1586), Vero (ATCC, CCL-81), BHK21 (ATCC, CCL-10) and 293 T (kindly gifted by Dr Jiahuai Han) cells were maintained in high-glucose DMEM (Sigma-Aldrich) supplemented with 10% FBS (Gibco), penicillin (100 IU/mL), and streptomycin (100 μg/mL) in a 5% CO2 environment at 37°C and passaged every 2 days. BHK21-hACE2 cells stably expressing hACE2 were developed by the PiggyBac transposon system. Transposon vectors (SBI System Biosciences, PB514B-2) containing the hACE2 gene and transposase plasmids were co-transfected into BHK21 cells and then selected through puromycin resistance and red fluorescence.
SARS-CoV-2 spike-specific antibody was generated through the immunization of Balb/c mice with SARS-CoV-2 spike protein (RBD, Fc Tag, 40592-V05H, Sino Biological Inc.). MAbs against the SARS-CoV-2 RBD were produced with hybridoma technology and were characterized by pseudotyped virus assay.
Twelve SARS-CoV-2 serum samples from convalescent patients were generously provided by Mr Zhi-Yong Li from the First Hospital of Xiamen University. Written informed consent was obtained from all volunteers.
Plasmids and pseudovirus
To construct a VSV pseudovirus carrying the spike protein of SARS-CoV-2, the spike gene of the Wuhan-Hu-1 strain (GenBank: MN908947) was codon-optimized for expression in human cells and cloned into the eukaryotic expression plasmid pCAG to generate the recombinant plasmid pCAG-nCoVS. The spike gene mutant of SARS-CoV-2 with a C-terminal 18 aa truncation was also cloned into plasmid pCAG and generated pCAG-nCoVSdel18. The plasmids pCAG-nCoVS and pCAG-nCoVSdel18 were each transfected into Vero-E6 cells. Forty-eight hours post transfection, the VSVdG-EGFP-G (Addgene, 31842) virus was inoculated into cells expressing the SARS-CoV-2 spike protein or the SARS-CoV-2 Sdel18 truncated protein and incubated for one hour[Citation15]. Then, VSVdG-EGFP-G virus was removed from the supernatant, and anti-VSV-G rat serum was added to block the infectivity of residual VSVdG-EGFP-G. The progeny virus would be enveloped by SARS-CoV-2 spike protein or SARS-CoV-2 Sdel18 truncated protein to generate a VSV pseudovirus carrying a different spike protein of SARS-CoV-2, as shown in . The supernatant was harvested at 24 h post VSVdG-EGFP-G infection and then centrifuged and filtered (0.45-μm pore size, Millipore, SLHP033RB) to remove cell debris and stored at −80°C until use. The viral titer was determined by counting the number of GFP-positive cells after BHK21-hACE2 cell infection with gradient diluted supernatant.
Pseudovirus-based neutralization assay
BHK21-hACE2 cells were seeded previously. The cultured supernatant of monoclonal hybridoma cells, gradient diluted purified antibodies or the serum of convalescent patients were mixed with diluted VSV-SARS-CoV-2-Sdel18 virus (MOI=0.05) and incubated at 37°C for 1 h. All samples and viruses were diluted with 10% FBS-DMEM. The mixture was added to seeded BHK21-hACE2 cells. After 12 h of incubation, fluorescence images were obtained with Opera Phenix or Operetta CLS equipment (PerkinElmer). For quantitative determination, fluorescence images were analyzed by the Columbus system (PerkinElmer), and the numbers of GFP-positive cells for each well were counted to represent infection performance. The reduction (%) in the number of GFP-positive cells in mAb-treated wells compared with that in nontreated control wells were calculated to show the neutralizing potency.
Wild type SARS-CoV-2-based neutralization assay
Vero cells were seeded 24 h before the infection in a 96-well plate (1 × 104cells/well). On the day of infection, the cells were washed twice. Serum samples were incubated at 56°C for 30 min and then diluted 2-fold in cell culture medium. Aliquots (50 μL) of diluted serum samples (from 20-fold to 10240-fold) were added to 50 μL of cell culture medium containing 50 times the tissue culture infective dose (TCID50) of wild type SARS-CoV-2 virus on a 96-well plate and incubated at 37°C for 2 h in CO2 5% vol/vol. The mixture of virus and serum were then added to cells in 96-well plates and plates were incubated at 37°C with microscopic examination for cytopathic effect (CPE) after a 5-day incubation. The highest dilution of serum that showed complete inhibition activity of SARS-CoV-2 was recorded as the neutralizing antibody titer (ID100). Assays were performed in duplicate with negative control serum.
Statistics
The inhibition rates of mAb and serum were calculated according to the decrease in the number of GFP-positive cells (for pseudotype-based neutralization assay) or the decrease in CPE (for wild type SARS-CoV-2-based neutralization assay). The IC50 (the half maximal inhibitory concentration) or ID50 (plasma/serum dilutions causing a 50% reduction in the number of GFP-positive cells) values were calculated with nonlinear regression, i.e. log (inhibitor) vs. response (four parameters), with GraphPad Prism 7.00 (GraphPad Software, Inc., San Diego, CA, USA). The correlation of the analysis results between the pseudovirus system and the wild type virus system was analyzed by linear regression with GraphPad Prism 7.00.
Results
The combination of VSV-SARS-CoV-2-Sdel18 and BHK21-hACE2 cells showed higher efficiency
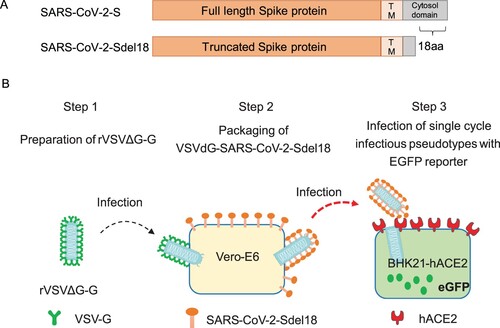
To compare the infection efficiency of pseudotyped viruses bearing different glycosylated proteins, single-cycle infectious recombinant VSVdG-EGFP-G was prepared and used to infect Vero-E6 cells transiently expressing full-length SARS-CoV-2 S protein and truncated SARS-CoV-2-Sdel18 protein with a C-terminal 18 aa truncation. Pseudotyped VSV-SARS-CoV-2-S and VSV-SARS-CoV-2-Sdel18 were obtained following the procedures described in the methods (). Vero-E6, BHK21, BHK21-hACE2 and 293 T cells were seeded to compare the infection efficiency of pseudotyped viruses. BHK21 cells stably expressing hACE2 (BHK21-hACE2 cells) were constructed as described in the methods, and the expression of hACE2 was confirmed (Supplementary figure 1). The infection efficiency of VSV pseudotypes bearing truncated Sdel18 was much higher than that of those bearing full-length S protein. Similar results were observed in BHK21-hACE2 and Vero-E6 cells, among which BHK21-hACE2 cells stably overexpressing hACE2 were most sensitive, whereas BHK21 cells and 293 T cells were almost resistant to infection (). The number of GFP-positive cells in VSV-SARS-CoV-2-Sdel18-infected BHK21-hACE2 cells were approximately 70 times greater than that in the VSV-SARS-CoV-2-S infection group and 1.26 times lower than that in the VSVdG-EGFP-G infection group (). Vero-E6 cells expressing C-terminal truncated spike protein (Sdel18) also formed more syncytia than cells expressing full-length spike protein (Supplementary figure 2). The results suggested that the combination of VSV-SARS-CoV-2-Sdel18 and BHK21-hACE2 cells could be a better alteration for establishing pseudotyped SARS-CoV-2 entry and neutralization assays.
Figure 2. Comparison of the infection efficiency of pseudotyped viruses in various cell lines. VSVdG viruses bearing the spike protein of SARS-CoV-2 or the G protein of VSV were harvested, and the infectivity of these recombinant viruses were tested in different cell lines, including Vero-E6, BHK21, 293 T and BHK21-hACE2 cells. The fluorescence was detected (A), and the numbers of GFP-positive cells (B) were counted with Opera Phenix 12 h post infection.
Vero-E6 cells packaged VSVdG-SARS-CoV-2-Sdel18 with the highest efficiency
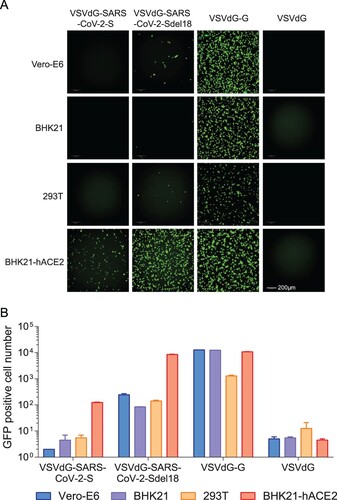
Viral packaging efficiency is one of the limiting factors for high-throughput in vitro neutralization assays. To select a cell line that is most appropriate for pseudovirus production, we compared the viral titers of VSVdG-SARS-CoV-2-Sdel18 packaged by Vero-E6, BHK21 and 293 T cells. The results showed that Vero-E6 cells formed more syncytia than the other two cell lines during packaging and produced the highest viral titer (), which was approximately 3 × 105 infectious particles per millilitre.
Figure 3. Comparison of the packaging efficiency of VSVdG-SARS-CoV-2-Sdel18 in various cell lines. Vero-E6, BHK21 and 293 T cells were used to package the VSVdG-SARS-CoV-2-Sdel18 virus. (A) The left picture shows the cells used to package recombinant virus, recorded 48 h post infection with VSVdG-EGFP-G. The right figures show the infectivity of virus produced by three cell lines. The harvested virus was diluted and tested in BHK21-hACE2 cells. The fluorescence was detected (A), and the numbers of GFP-positive cells (B) were counted with Opera Phenix 12 h post infection.
Time course of EGFP expression after VSVdG-SARS-CoV-2-Sdel18 infection
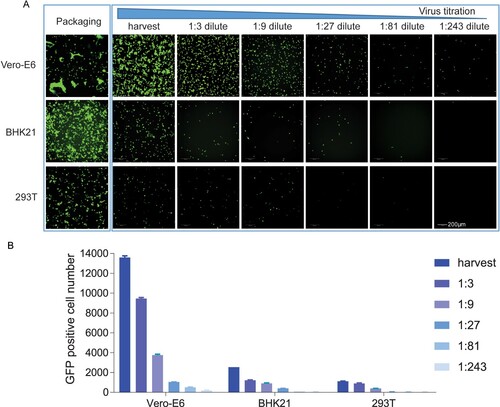
To determine the optimal incubation period for the detection of VSVdG-SARS-CoV-2-Sdel18 infection, time-course monitoring of EGFP expression was performed with the Opera Phenix High Content Screening System. EGFP expression could be detected as early as 8 h post infection, and the number of GFP-positive cells gradually increased, reaching the plateau phase within 12 h, and remained stable between 12 and 24 h (). However, the fluorescence intensity increased continuously. As pseudoviruses are unable to produce progeny viruses, the infected cell number would be a better indicator for infection efficiency. Therefore, the infection efficiency could be quantitatively evaluated at 12–24 h by counting the number of GFP-positive cells.
Figure 4. Time course of EGFP expression after VSVdG-SARS-CoV-2-Sdel18 infection. BHK21-hACE2 cells were infected with VSVdG-SARS-CoV-2-Sdel18 virus (MOI=0.05). The fluorescence was detected (A), and the numbers of GFP-positive cells (B) were counted with Opera Phenix at different time points post infection.
Optimization and validation of the neutralization assay
To optimize the number of cells seeded, a range of 6.25 × 103–2.00 × 105 cells/well were seeded and infected with an inoculant dose of 0.05 MOI. The ID50 values were calculated with nonlinear regression, i.e. log (inhibitor) vs. response (four parameters), and the goodness of fit test showed that R2 was greater than 0.98, revealing an excellent fitting of the four-parameter inhibition curves. The ID50 values were similar, with cell numbers ranging from 6.25 × 103 to 5.00 × 104 cells/well (A). When the number of cells seeded was more than 5.00 × 104 cells/well, the ID50 values tended to decrease. Given these results, 5.00 × 104 cells/well was selected for subsequent experiments.
Figure 5. Optimization in the number of seeded cells and inoculant dose of pseudovirus (A) To optimize the seeding cell number, a range of 6.25 × 103–2.00 × 105 cells/well were seeded and infected with the inoculant dose of 0.05 MOI, the ID50 values were calculated with non-linear regression, i.e. log(inhibitor) vs. response (four parameters). (B) The optimal inoculant dose of pseudovirus was tested with a range from 0.00625 to 0.2 MOI.
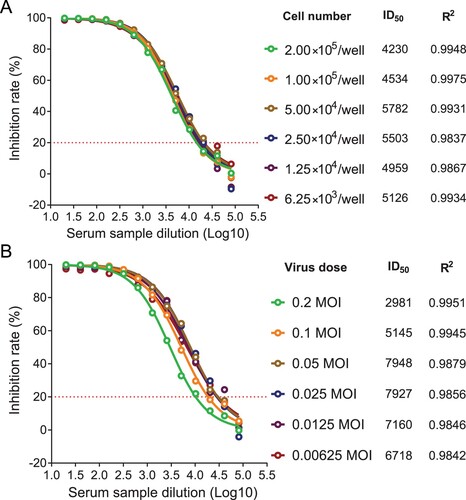
Furthermore, the optimal inoculant dose of pseudovirus was tested with a range from 0.00625 to 0.2 MOI. Typical four-parameter inhibition curves were observed, and the R2 values were greater than 0.98 in all groups, revealing an excellent fitting of nonlinear regression curves. The ID50 values were similar with inoculant doses ranging from 0.0125 to 0.05 MOI (B), and when the MOI was more than 0.05, the ID50 values tended to decrease. According to these results, 0.05 MOI was used as the optimal inoculant dose.
To determine the cutoff value of the assay, a negative sample panel including 59 human sera and 58 mouse sera were used, with an initial dilution of 1:10 followed by a 3-fold serial dilution, and then the 50% inhibitory dilution (ID50) was calculated. The background (mean ± SD) of negative serum samples from humans and mice were 1.97 ± 1.96 and 9.86 ± 5.91, respectively (A). The lower limit of detection (LLOD) was calculated by the mean+1.96 SD, and the LLOD was 5.82 for human serum samples and 21.45 for mouse serum samples. The cutoff values were set as 10 and 30 for human and mouse serum samples, respectively.
Figure 6. Validation of the VSVdG-SARS-CoV-2-Sdel18 pseudovirus assay. (A) Specificity of the pseudovirus assay. A negative sample panel including 59 human sera and 58 mouse sera were used to determine the specificity of this assay. (B) Reproducibility of the pseudovirus assay. One COVID-19 convalescent patient serum sample was tested 14 times on individual plates in three independent experiments. The virus titer of VSVdG-SARS-CoV-2-Sdel18 pseudovirus was consistent in these assays (MOI=0.05). (C) The correlation of neutralizing titer measured by the VSVdG-SARS-CoV-2-Sdel18 pseudovirus assay (ID50, log10) and the wild type SARS-CoV-2 neutralization assay (ID100, log10).
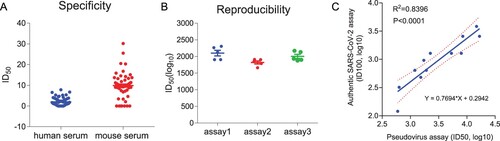
To test the reproducibility of this neutralization assay, one COVID-19 convalescent patient serum sample was tested 14 times on individual plates in three independent experiments, and the average intra-assay and inter-assay coefficient of variation (CV) were 7.2% and 9.2%, respectively (B), which was acceptable for a cell-based assay (CV≤25%).
To validate the correlation of neutralizing antibody titers measured by our VSVdG-SARS-CoV-2-Sdel18 pseudovirus assay and wild type SARS-CoV-2 assay, a total of 12 serum samples from COVID-19 convalescent patients were used for quantitative measurement. Viral infection was determined by counting GFP-positive cells (pseudovirus) or examining CPE (authentic virus). For the pseudovirus-based neutralization assay, ID50 was calculated with the quantitative reduction of the number of GFP-positive cells. For the wild type SARS-CoV-2 neutralization assay, the neutralizing antibody titers were determined by microscopic examination for CPE and the highest dilution of serum that showed complete inhibition (ID100) of CPE was used to indicate the neutralizing antibody titer in serum. The results showed that the neutralizing titers measured by the two assays were highly correlated, with an R squared value of 0.8396 (P<0.0001) (C). The geometric mean neutralizing titers (GMT) measured by the pseudovirus assay and wild type SARS-Cov-2 assay were 2961 and 923, respectively (Supplementary Table 1). It should be noted that the neutralization titers measured by assays from different laboratories are different, and the same serum plate needs to be used for calibration before they can be compared with each other.
VSVdG-SARS-CoV-2-Sdel18-based system for the screening of neutralizing mAbs
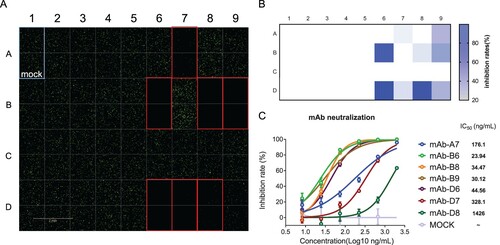
Thirty-five strains of mouse monoclonal antibody hybridoma cultured supernatants were detected, and 7 of them showed significant neutralizing activity (A, B). The IC50 of the 7 selected neutralizing antibodies for antiviral activity were also measured (C). The results suggested that this system can be used for high-throughput screening of neutralizing antibodies.
Figure 7. VSVdG-SARS-CoV-2-Sdel18-based neutralization assay for screening neutralizing mAbs. (A) Measurement of the neutralizing activity of 35 strains of antibodies. The cultured supernatant of 35 monoclonal hybridoma cells were incubated with VSVdG-SARS-CoV-2-Sdel18 virus (MOI=0.05), and then the mixture was added to BHK21-hACE2 cells. The fluorescence was detected with Opera Phenix 12 h post infection. (B) The numbers of GFP-positive cells were counted to calculate the inhibition rate. (C) The IC50 values of the 7 selected neutralizing antibodies for antiviral activity were also analyzed. The 7 selected neutralizing antibodies were purified and diluted to different concentrations, incubated with VSVdG-SARS-CoV-2-Sdel18 virus (MOI=0.05) for an hour and added to BHK21-hACE2 cells. The numbers of GFP-positive cells were counted with Opera Phenix 12 h post infection to calculate the inhibition ratio. The IC50 was analyzed by nonlinear regression (four-parameter).
Discussion
The COVID-19 epidemic is spreading rapidly around the world, and there is an urgent need for vaccines or therapeutic drugs. The isolation and culture of SARS-CoV-2 have to be handled in biosafety level-3 (BSL-3) facilities, which limits the research and development of drugs. A safer and more efficient assay for drug evaluation would accelerate the development of drugs.
The pseudovirus system provides a convenient tool for the study of RG-3 viruses, making the evaluation of viral inhibitors, neutralizing antibodies and immunized serum much easier. Retrovirus and VSV are both commonly used pseudovirus systems, but the pseudotype viral titer obtained with the VSVdG system is generally higher than that of the retrovirus system [Citation10]. Furthermore, the infection of target cells with pseudotyped VSV can be detected as early as 4 h post infection. In this research, pseudotyped VSV carrying the spike protein of SARS-CoV-2 and a reporter gene can be detected as early as 8 h post infection through green fluorescence. This time is much shorter than the time to detect wild type SARS-CoV-2. The experimental results can be obtained by automatically counting the number of EGFP-positive cells at 12 h after infection, making the assay convenient and high-throughput.
Jianhui Nie et al. constructed a pseudotyped VSV bearing the full-length SARS-CoV-2 S protein [Citation12]. Notably, our comparative experiments showed that truncating the 18 amino acids of the C-terminal cytoplasmic tail of the SARS-CoV-2 S protein could significantly improve the infection efficiency of this system (Supplementary figure 3). This finding is similar to that of a recently published replication-competent rVSV-SARS-CoV-2-S study, which found that rVSV-SARS-CoV-2-S replicated poorly initially, and accelerated growth was observed after 5 passages, which coincided with the emergence of mutants with 21- or 24-amino acid truncations of the cytoplasmic tail of the S protein [Citation16]. The C termini of the SARS-CoV-2 S protein and the SARS-CoV S protein are very similar, and both carry signals for endoplasmic reticulum positioning. The truncation of 18–19 amino acids has been found to increase infection efficiency [Citation6,Citation11,Citation14].
The infection model consisting of BHK21-hACE2 cells and VSVdG-SARS-CoV-2-Sdel18 can mimic the entry of wild type SARS-CoV-2. The serum neutralizing titer of COVID-19 convalescent patients measured by the VSV-SARS-CoV-2-Sdel18 pseudovirus assay has a good correlation with that measured by the wild type SARS-CoV-2 assay. The pseudovirus assay is safer and more time saving than the wild type SARS-CoV-2 assay.
This pseudotype infection assay could be applied to the screening of compounds that could inhibit infection by SARS-CoV-2 and neutralizing antibodies. More importantly, this method can be used to measure the neutralization titer of serum from vaccinated animals or humans, which is fast and convenient. Taken together, this neutralization assay based on single-cycle infectious VSV-SARS-CoV-2 Sdel18 pseudovirus and BHK21-hACE2 cells could greatly benefit the development of vaccines and drugs against COVID-19.
Author contribution
Hua-Long Xiong, Bao-Ying Huang, Zhi-Yong Li, Tian-Ying Zhang, and Ning-Shao Xia had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Tian-Ying Zhang, Quan Yuan and Ning-Shao Xia. Acquisition of data: Hua-Long Xiong, Yang-Tao Wu, Jia-Li Cao, Jian Ma, Xiao-Yang Qiao, Bao-Hui Zhang, Yang-Shi, Jing-Jing Xu, Liang-Zhang. Ying-Xia Liu, Ren Yang and Bao-Ying Huang performed the authentic virus assay. Analysis and interpretation of data: Hua-Long Xiong, Jia-Li Cao, Tian-Ying Zhang. Drafting of the manuscript: Tian-Ying Zhang, Jia-li Cao, Hua-Long Xiong. Critical revision of the manuscript for important intellectual content: Sheng-Xiang Ge, Jun Zhang, Quan Yuan, Tian-Ying Zhang, and Ning-Shao Xia. Technical or material support: Shao-Juan Wang, Wang-Heng Hou, Ya-Li Zhang, Bao-Rong Fu, Xiang-Yang Yao, Zhi-Yong Li and Ting Yang. Study supervision: Bao-Ying Huang, Zhi-Yong Li, Tian-Ying Zhang and Ning-Shao Xia.
Ethics approval
Experiments using human serum were approved by the Medical ethics committee of School of Public Health of Xiamen University. The experiments in mice were conducted under approval of the Institutional Animal Care and Use Committee at Xiamen University and were in accordance with the Guide for the Care and Use of Laboratory Animals.
Supplementary.docx
Download ()Acknowledgements
We sincerely thanks for the team of Prof. Yong-Zhen Zhang, who publishing the genome sequence of SARS-CoV-2 (GenBank: MN908947) in the early stage of the epidemic. This work was supported by the National Natural Science Foundation of China (81993149041, 81702006), Science and Technology Major Project of the Fujian Province (2020YZ014001), Xiamen Science and Technology Major Project (3502Z2020YJ02) and National Key Research and Development Program of China (2016YFD0500301).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article was originally published with errors, which have now been corrected in the online version. Please see Correction https://doi.org/10.1080/22221751.2024.2324415
Additional information
Funding
References
- Phelan AL, Katz R, Gostin LO. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020;323.8:709–710. doi: 10.1001/jama.2020.1097
- Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316
- Hui DS, I Azhar E, Madani TA, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8
- Hofmann H, Hattermann K, Marzi A, et al. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004
- Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11.1:1–12.
- Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181.2:271–280.e8. doi: 10.1016/j.cell.2020.02.052
- Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762
- Ogino M, Ebihara H, Lee B-H, et al. Use of vesicular stomatitis virus pseudotypes bearing hantaan or Seoul virus envelope proteins in a rapid and safe neutralization test. Clin Diagn Lab Immunol. 2003;10:154–160.
- Fukushi S, Mizutani T, Saijo M, et al. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J Gen Virol. 2005;86:2269–2274.
- Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9:680–686.
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569.
- Schwegmann-Weßels C, Glende J, Ren X, et al. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J Gen Virol. 2009;90:1724–1729.
- Whitt MA. Generation of VSV pseudotypes using recombinant DeltaG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods. 2010;169:365–374.
- Dieterle ME, Haslwanter D, Bortz RH, et al. A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 spike-mediated cell entry and its inhibition. Cell Host Microbe. 2020;28:1–11.