ABSTRACT
HBV cccDNA stably exists in the nuclei of infected cells as an episomal munichromosome which is responsible for viral persistence and failure of current antiviral treatments. However, the regulatory mechanism of cccDNA transcription by viral and host cellular factors is not well understood. In this study, we investigated whether cccDNA could be recruited into a specific region of the nucleus via specific interaction with a cellular chromatin to regulate its transcription activity. To investigate this hypothesis, we used chromosome conformation capture (3C) technology to search for the potential interaction of cccDNA and cellular chromatin through rcccDNA transfection in hepatoma cells and found that cccDNA is specifically associated with human chromosome 19p13.11 region, which contains a highly active enhancer element. We also confirmed that cellular transcription factor Yin-Yang 1 (YY1) and viral protein HBx mediated the spatial regulation of HBV cccDNA transcription by 19p13.11 enhancer. Thus, These findings indicate that YY1 and HBx mediate the recruitment of HBV cccDNA minichromosomes to 19p13.11 region for transcription activation, and YY1 may present as a novel therapeutic target against HBV infection.
Introduction
Hepatitis B virus (HBV) is the major causative agent of chronic hepatitis B (CHB), cirrhosis and hepatocellular carcinoma. Currently, 257 million people are chronically infected by HBV, and 887,000 people die from HBV infection-related liver diseases annually [Citation1, Citation2]. During HBV replication, the converted into covalently closed circular (ccc) DNA of HBV serves as the template for transcription of viral RNAs, which can be assembled into an episomal minichromosome in the nuclei of infected hepatocytes by hijacking host factors. It has been reported that the pre-existing cccDNA reservoir is responsible for viral persistence. The failure of curing the CHB by the current standard of care medication is primarily due to the lack of direct effects against cccDNA metabolism and function [Citation3]. As a result, viral replication rebound usually occurs upon the discontinuation of antiviral therapy. Therefore, elimination or functional inactivation of cccDNA from infected hepatocytes, to achieve viral clearance, has become a major issue in the cure of chronic hepatitis B (CHB).
HBV has a complicated, but unique life cycle. The infection of HBV in hepatocyte begins from binding to the sodium taurocholate co-transporting polypeptide (NTCP) receptor and delivering its nucleocapsid into the cytoplasm. The relaxed circular DNA (rcDNA) genome is then transported into the nucleus and formed cccDNA. The cccDNA genome contains four coding genes(S, P, C and X), four promoters (CP, SP I, SP II, and XP) and two enhancers (EN I and EN II) [Citation4]. Though employing the cellular transcription machinery, cccDNA acts as the transcriptional template for viral mRNA and pregenomic RNA(pgRNA). Upon translation of viral proteins, pgRNA and DNA polymerase are packaged into nucleocapsids where reverse transcriptional viral DNA synthesis takes place. The progeny cytoplasmic nucleocapsids are subsequently enveloped and secreted out of the hepatocytes as infectious virions [Citation5]. As we know, the cccDNA associates with nucleosomes and non-histone proteins and exists as an episomal minichromosome. Viral HBx and core proteins have also been demonstrated to be recruited to cccDNA minichromosome and regulate its transcription activity [Citation6]. Additionally, similar to host cellular chromatin, cccDNA minichromosome can be epigenetically modified to regulate viral genes expression, including DNA methylation and post-translational histone modifications, such as methylation and acetylation [Citation7–9]. However, the regulatory mechanism of cccDNA transcription by viral and host cellular factors is not fully understood.
In the nucleus, both intrachromosomal and interchromosomal long-range associations have been demonstrated to bring widely separated functional DNA elements into close spatial proximity and create interactions between these elements [Citation10, Citation11]. For instance, the expression of human neuroglobin gene is controlled by a distal regulatory element located −70 kb upstream from the gene in neuroblastoma cells [Citation12]. Our previous study also demonstrated a long-range chromatin interaction between integrated HPV DNA fragments, the proto-oncogene MYC, and the 8q24.22 chromosomal region in HeLa cells [Citation13]. The interaction between chromosome elements requires one or multiple cellular proteins, such as CTCF, cohesion and Yin-Yang 1 (YY1) [Citation14–16].
As an episomal minichromosome, we speculated whether cccDNA in nucleus can be recruited to specific nuclear regions. Currently, two studies had demonstrated that HBV cccDNA is associated with highly active transcription sites on host chromosomes [Citation17, Citation18], but the role of one specific host chromatin on HBV cccDNA has never been mentioned or analysed. In the present study, we are interested in determining whether cccDNA can make interaction with a cellular chromatin to regulate its transcriptional activity. Interestingly, we identified that cccDNA specifically associated with chromosome 19p13.11 region, which contains a highly active enhancer element, leading to cccDNA transcription activation mediated by YY1 and viral protein HBx. Thus, our findings provide new insights into the mechanism by which the spatial interaction of cccDNA minichromosome with cellular chromosome promotes cccDNA transcription and HBV replication.
Material and methods
Cell culture
Huh7 and HepG2 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured by Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 μg/ml streptomycin (Gibco, Carlsbad, CA, USA). Cells were placed at 37°C in 5% CO2 humidified incubators.
Plamids and siRNA
PrcccDNA and pCMV-Cre plasmids were provided by Professor. Deng Qiang at Fudan University, Shanghai, China. A loxP-chimeric intron was engineered into a monomeric HBV genome in a precursor plasmid (prcccDNA), which was excised using Cre/loxP-mediated DNA recombinationin to a 3.3-kb rcccDNA in the nuclei of hepatocytes. In cultured hepatoma cells, cotransfection of prcccDNA and pCMV-Cre (en-coding Cre recombinase) resulted in accumulation of nuclear rcccDNA that was heat stable and epigenetically organized as a minichromosome [Citation19, Citation20]. The siRNA-YY1 and siRNA-mock were obtained from Guangzhou RiboBio Co., Ltd (Ribobio, GuangDong, China). The HBx-mutated prcccDNA plasmid without HBx protein expression was constructed from C base to T base at HBV 1397nt.
Chromosome conformation capture (3c)
PrcccDNA and pCMV-Cre plasmids were co-transfected at a molar ratio of 1:1 into Huh7 or HepG2 cells using lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA). The cells were harvested at 3 days post transfection for 3C analyses by following the published procedure [Citation21]. Chromosomal DNA and mini-chromosomal HBV cccDNA were fragmented by TaqI restriction enzyme (NEB, Ipswich, MA, USA) digestion at 65°C for 16 h, and inactivated by incubation at 80°C for 30 min. The DNA fragments were ligated by T4 DNA ligase (NEB, Ipswich, MA, USA) and extracted by phenol–chloroform extraction. Finally, PCR amplification were performed by combined use of primers specific for HBV and those specific for, human chromosome Alu and or 19p13.11 sequences. ACTB DNA was used as a loading control for the 3C assay. HBV-ACTB ligation was used as a negative control for the specificity of the assay. The sequences of primers used in 3C assays are shown in supporting information Table S1.
Chromatin immunoprecipitation (ChIP)
Huh7 or HepG2 cells were co-transfected with plasmids prcccDNA and pCMV-Cre at a molar ratio of 1:1 and harvested at 3 days post transfection. Chip assays were performed by using SimpleChIP® Enzymatic Chromatin IP Kit (CST, Danvers, MA, USA) according to the manufacturer's instructions. PCR assays were performed by using primers specific to HBV or 19p13.11 sequences, respectively. The sequences of primers used in Chip assays are shown in supporting information Table S1.
Establishment of 19p13.11-ko Huh-7 and HepG2 cells
LentiCRISPR V2 plasmid was digested by BsmBI enzyme (NEB, Ipswich, MA, USA) and constructed with gRNA-19p13.11. gRNA sequences targeted 5′ or 3′ end of 19p13.11 enhancer was shown in supporting information Table S1. Supernatant was collected from 293FT cells transfected LentiCRISPR V2 plasmids together with packaged plasmids 2 days later and added into Huh7 or HepG2 cells in culture. Cells were continuingly cultured with 1 mg/ml puromycin for another two weeks, and the positive clones that 19p13.11 enhancer region was knock out were identified by PCR assays.
Cell counting with CCK8 kit
2×103 cells were cultured with 200μl medium in a well of one 96 well plate, and the wild or 19p13.11-ko cells of Huh7 and HepG2 were seeded in quintuple, respectively. According to the instruction manual of cell counting kit (cck-8) (Solarbio, Beijing, China), 20μl cck-8 solution was added into each well with cells, and incubated for another hour at 37°C in the 5% CO2 humidified incubator. Then the absorbance value was detected under the wavelength of 450 nm by use of iMark enzyme standard instrument (Bio-Rad, Hercules, CA, USA).
Detection of HBsAg, HBeAg and HBV total RNAs
The supernatant and cells co-transfected by prcccDNA and pCMV-Cre plasmids (1:1 in ratio) were collected 3 days later. For the supernatant, the levels of HBsAg and HBeAg were measured by the diagnostic kit for the quantitative determination of HBsAg or HBeAg (time-resolved Immunofluorometric Assay) (PerkinElmer, Waltham, MA, USA) following the manufacturer's instructions. For the cells, Total RNAs were extracted by trizol. And RNAs were reverse-transcribed using First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland) with random primers. Then the expression level of HBV total RNAs was measured by specific primers designed by our previous work [Citation22].
Engineered DNA binding molecule mediated chromatin immunoprecipitation (enChIP)
PrcccDNA, pCMV-Cre and px330-gRNA-HBV-Flag-dCas9 plasmids (Addgene, Cambridge, MA, USA) were co-transfected into Huh7 cells, and enChip assays were developed 3 days later according to the literature [Citation23] and the above ChIP protocol. Briefly, DNAs-proteins were cross-linked by 1% formaldehyde and stopped by 127 nm Glycine solution. Then the nuclear pellets were collected by lysis buffer, and chromatin was sonicated by ultrasound. Next, Anti-Flag antibody (Sigma-Aldrich F1804, 1:50, St. Louis, MO, USA) was added to the above contents and incubated at 4°C for overnight. Proteins were received by adding appropriate amount of sample buffer to the beads together with incubating at 100°C for 30 min, and DNAs were eluted by adding chip elution buffer to the beads and received by phenol chloroform extraction. Finally, proteins were detected by anti-cas9 (CST 65832, 1:1000) or anti-YY1 antibody (CST 63227, 1:1000, Danvers, MA, USA) through western blots, and DNAs were measured through PCR by the same primers of HBV and 19p13.11 in Chip assays.
Co-IP experiments
pCMV-vector was bought from Beyotime technology (Shanghai, China). YY1 (HA tag in N terminal) and HBx (His tag in N terminal) sequences were constructed into pCMV-vectors through KpnI and XbaI enzyme (NEB, Ipswich, MA, USA) digestion. PCMV-HA-YY1 and pCMV-His-HBx vectors were co-transfected into Huh7 cells and co-IP assays were proceeded 2 days later. Briefly, 500μl cell lysis buffer (Beyotime, Shanghai, China) was used and then 1:50 diluted anti-HA (CST 3724, Danvers, MA, USA) or anti-His antibody (CST 12698, Danvers, MA, USA) was added into the solution at 4°C for overnight. Then magnetic beads were added and incubated for another 2 h. Later, beads were collected by magnetic separation rack and washed by lysis buffer mentioned above for five times. And finally, proteins were received by adding sample buffer to the beads with blending and incubating at 100°C for 30 min. The collected proteins were detected through western blots by anti-YY1 (CST 63227, Danvers, MA, USA) or anti-HBx antibody (Abcam ab2741, Cambridge, UK) in a concentration of 1:1000 dilution.
Statistical analyses
The difference of relative expression between two groups was analysed by t test in GraphPad and the P value <0.05 was considered to be significant.
Results
HBV cccDNA in nucleus is close to chromosome 19p13.11 enhancer locus in term of spatial proximity
To produce enough rcccDNA, we co-transfected prcccDNA and pCMV-Cre plasmids into Huh7 cells which could induce the accumulation of nuclear rcccDNA that was heat stable and epigenetically organized as a minichromosome [Citation19, Citation20]. Then we performed Chromosome conformation capture (3C) technique to identify the potential human chromosome regions close to cccDNA spatially. The schematic diagram of 3C procedure was shown in (A) [Citation21]. HBV genome specific primer in combination with primer homologous to the highly frequently repeated Alu sequences in human genome was used to detect viral-host junctions [Citation24]. If HBV cccDNA has close spatial proximity with a cellular chromosome region containing Alu sequence, the PCR assay of 3C would amplify the corresponding cross-linked DNAs as PCR template. As shown in (B), we obtained a unique PCR product in crosslinked cells by using primer pair of HBV F and Alu R. The subsequent sequencing analysis revealed that this product was a fusion fragment consisted of partial HBV and partial human genome sequences mediated by TaqI enzyme digestion sites. When we searched the database of human genome (http://genome.ucsc.edu) for sequence homologous to this fragment, it came out that this fragment mapped to chromosome 19p13.11 region.
Figure 1. Physical proximity of 19p13.11 region and HBV cccDNA detected by 3C assays. A: Schematic diagram of 3C procedure. B: HBV-Alu ligation fragments detected by 3C assays. The PCR products were obtained from cross-linked Huh7 cells with specific pairs of primers of HBV and Alu sequence: HBV F + Alu F, HBV R + Alu F, HBV F + Alu R, HBV R + Alu R. Among them, a unique PCR product was shown by using HBV F + Alu R primers. C: 19p13.11-HBV ligation fragments detected by 3C assays. The PCR products were obtained from cross-linked Huh7 and HepG2 cells respectively with a specific pair of primers of 19p13.11 and HBV sequence: 19p13.11 R + HBV F (540bp) (upper). The sequencing of these fragments identified 19p13.11 – HBV piecing sequences mediated by TaqI enzyme site (TCGA) (lower). HBV + ACTB (350bp) was used as a specificity control, and ACTB gene (288 bp) was used as a loading control.
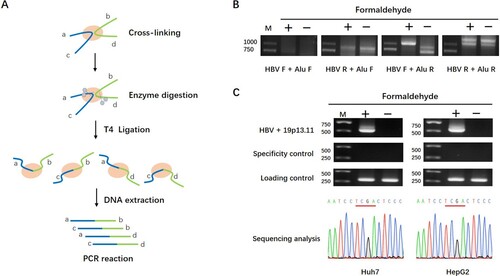
Next, to confirm the close co-location in spatial structure between HBV genome and chromosome 19p13.11 region, primer specific to 19p13.11 region was designed in pair with the HBV specific primer F. Again, a unique PCR product was obtained in the crosslinked cells, and subsequent sequence analysis of the PCR product confirmed the close spatial proximity of HBV cccDNA with cellular chromosome 19p13.11 region in Huh7 and HepG2 cells, respectively (C). Moreover, the contamination of prcccDNA could be simply excluded by the size of 3C-PCR products because of the unique position of HBV primer F on HBV genome (Fig S1A). Taken together, these results indicated that the HBV cccDNA genome in nucleus located quite close to 19p13.11 region in spatial distance.
The enhancer sequence in 19p13.11 region could spatially regulate the transcriptional activity of HBV cccDNA
Based on the annotation of database (http://genome.ucsc.edu), we found that there is no protein-coding gene in or near (<10 kb) this 19p13.11 region. However, a 7106 bp long potential enhancer element (GeneCards: GH19J018966) is identified in this region, which has high intensity of H3K4me1 and H3K27ac modification (A). As we know, intensive H3K27ac binding represents active enhancer activity [Citation25]. Indeed, through ChIP assays, we confirmed the obvious H3K27ac binding in this enhancer region in both HepG2 and Huh7 cell lines originally derived from human liver cancer, suggested that this region is a real active enhancer element in hepatocyte, or at least in malignant transformed hepatocyte (B).
Figure 2. 19p13.11 enhancer could regulate the transcriptional activity of HBV cccDNA. A: Analysis of 19p13.11 enhancer in UCSC database. Scale: the length of the enhancer, 7106bp. Chr19: the location of the enhancer, from 18966372 to 18973477 in hg38. Layered H3K4me1, H3K4me3, H3K27ac: the intensity of H3K4me1, H3K4me3, H3K27ac modification of the enhancer in seven cell lines distinguished by different colours including GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK and NHLF cells. The high intensity of H3K4me1 and H3K27ac were mostly shown in HSMM, HUVEC, K562, NHEK and NHLF cells. GH19J018966: the ID card of the regulatory element in GeneCards database and colours are used to distinguish promoters and enhancers. Red: promoters, Grey: enhancers. DNase clusters: clusters of DNaseI hypersensitivity derived from assays in 95 cell types. Regulatory regions in general, and promoters in particular, tend to be DNase-sensitive. B: Chip-PCR shows obvious H3K27ac binding in 19p13.11 site in Huh7 and HepG2 liver cancer cells. The intended PCR product is 228bp. C: The value of CCK8 absorbance of wild type or 19p13.11-ko Huh7 and HepG2 cells from first to fifth day. Three independent experiments were performed for each assay. D: The relative expression of HBsAg and HBeAg in supernatant detected by ELISA and HBV total RNAs intracellular detected by q-PCR of wild type or 19p13.11-ko Huh7 and HepG2 cells co-transfecting prcccDNA and pCMV-Cre plasmids three days later. Three independent experiments were performed for each assay. * P<0.05. ** P<0.01.
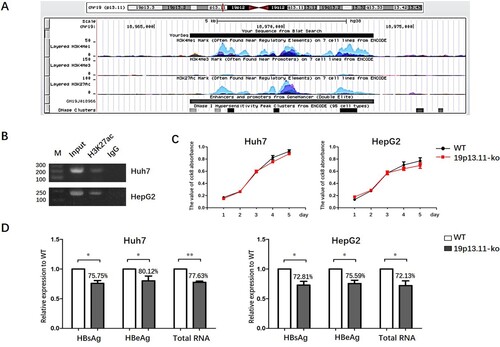
Next, we wondered whether this enhancer is involved in the regulation of HBV cccDNA transcription activity via spatial interaction. To address this question, firstly two gRNA sequences respectively targeting the 5′ and the 3′ ends of 19p13.11 enhancer were designed by use of Lentivirus-CRISPR-Cas9 system. And then, using Huh7 and HepG2 cell lines, we established 19p13.11 enhancer stably knockout cell strains (19p13.11-ko cell) (Figure S1B). We demonstrated that the proliferation rate of these 19p13.11-ko cells showed no statistical difference compared with each's wild type cells (C). Then, prcccDNA and pCMV-Cre plasmids were co-transfected into the wild type as well as 19p13.11-ko Huh7 and HepG2 cells and the intracellular HBV total RNA level, as well as the abundance of HBsAg and HBeAg in supernatant, were quantitatively measured 3 days post co-transfection. The results showed that HBV total RNA was reduced to 77.63% in 19p13.11-ko Huh7 cells compared with the wild type counterpart(P<0.01). Consistently, the expression of HBsAg and HBeAg in 19p13.11-ko Huh7 cells was decreased to 75.75%, and 80.12% (P<0.05, P<0.05), respectively. Similar results were also observed in HepG2 cells (D). In addition, to establish the specificity that only cccDNA can be recruited to 19p13.11, not a plasmid linked to a small part of the HBV genome, we performed such additional experiments with transfecting pcDNA3.1-precore plasmid (including HBV genome from 1814nt to 2452nt) or pcDNA3.1-HBs plasmid (including HBV genome from 2848nt to 835nt) into wild type or 19p13.11-ko Huh7 and HepG2 cells respectively. And the abundance of HBeAg in supernatant as well as intracellular L-HBsAg were quantitatively measured two days post transfection. As expected, the expression of HBeAg or L-HBsAg was not affected by 19p13.11 knock out in both Huh7 and HepG2 cells (p=0.32, p=0.71) (Figure S1C), Which further confirmed the specific interaction between 19p13.11 and HBV cccDNA. These results all indicated that 19p13.11 enhancer might play an important role in regulating the transcriptional activity of HBV cccDNA.
YY1 could combine with 19p13.11 and HBV genome and regulate HBV cccDNA transcription
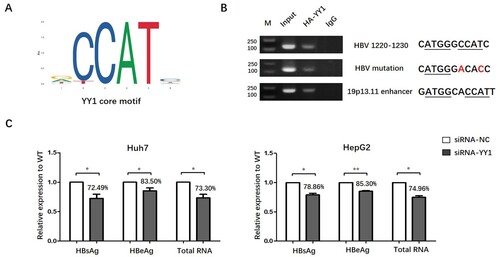
Previous studies have reported that cellular proteins including CTCF, cohesion and YY1 were involved in the spatial interaction between different chromosome elements [Citation14–16]. Therefore, we searched the potential binding motif of these cellular proteins in HBV genome as well as enhancer element at chromosome 19p13.11. We identified the presence of YY1 potential core binding motif (http://jaspar.genereg.net/) in both HBV genome (genotype D, NC_003977 in Genbank) and the 19p13.11 enhancer element, implicated that transcription factor YY1 might have the capability to bind to each of them (A). Then, we randomly selected 30 sequences of HBV from A-I genotypes and found that the YY1 binding site was pretty conservative (supporting information Table S2) in HBV genome. Next, ChIP-PCR experiments confirmed the binding of YY1 with 19p13.11 enhancer site and HBV genome. To further confirm that the binding of YY1 to HBV DNA is sequence dependent, the potential binding site of YY1 was mutated without affecting the amino acids of P protein in HBV genome 1220–1230nt. As expected, the binding of YY1 binding to HBV DNA was significantly decreased when the potential binding site of YY1 was mutated. Importantly, the expression of HBsAg, HBeAg and total RNA was reduced to 72.49%, 83.50% and 73.30% (P<0.05, P<0.05, P<0.05), respectively, when YY1 was knocked down in Huh7 cells. Similarly, the expression of HBsAg, HBeAg and total RNA in siRNA-YY1 HepG2 cells reduced to 78.86%, 85.30% and 74.96% (P<0.05, P<0.01, P<0.05), as compared to that in HepG2 cells with siRNA-mock (C).
Figure 3. YY1 could combine with 19p13.11 enhancer as well as HBV genome and regulate the transcriptional activity of HBV cccDNA. A: core motif of YY1 binding sites from JASPAR database. B: ChIP-PCR by anti-HA antibody shows obvious YY1 binding in potential 19p13.11 site (189bp) and HBV wild or mutated genome (130bp) in Huh7 cell line. Nucleotides underlined present core motif of YY1 binding and nucleotides marked red present mutation. C: The relative expression of HBsAg and HBeAg in supernatant detected by ELISA and HBV total RNAs intracellular detected by q-PCR of siRNA-mock or siRNA-YY1 as well as prcccDNA and pCMVCre plasmids transfected Huh7 and HepG2 cells 48 h later. Three independent experiments were performed for each assay. * P<0.05. ** P<0.01.
YY1 protein mediated the spatial interaction between 19p13.11 enhancer and HBV genome
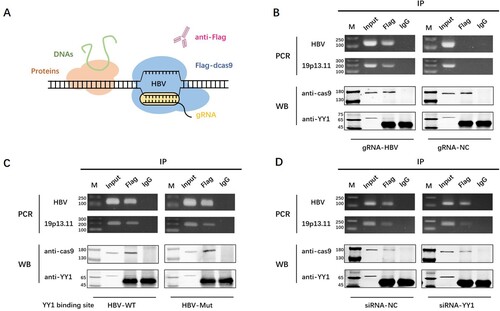
To further confirm whether YY1 protein mediated the spatial interaction between 19p13.11 enhancer and HBV genome, another technique called engineered DNA binding molecule mediated chromatin immunoprecipitation (enChIP) was performed [Citation23]. The schematic diagram of enChIP procedure was shown in (A). In the form of enChIP, genomic regions are immunoprecipitated with anti-Flag antibody against the Flag tag fused to dCas9 protein, which is co-expressed with a gRNA to recognize endogenous DNA sequence of interest in the genomic regions. Considering that the gRNA-targeted site cannot be too far away from the binding site of YY1 on HBV, we used a gRNA targeted to HBV 1180–1199nt which has been proved to identify HBV effectively and specifically in our previous work [Citation26]. Meanwhile, a gRNA targeted no sequence of human or HBV genome was used as negative control. As expected, we discovered the existence of HBV and 19p13.11 DNA sequence, together with dCas9 and YY1 protein in the same pull-down sample in Huh7 cells (B). Moreover, the abundance of 19p13.11 DNA was obviously decreased, along with the reduced YY1 binding by core binding motif mutation (C) or siRNA mediated expression reduction (D). These results indicated that YY1 might act as a structural and functional regulator to mediate the spatial proximity and interaction between 19p13.11 enhancer and HBV genome. Furthermore, we knocked down the expression of YY1 in 19p13.11-ko Huh7 and HepG2 cells, as well as transfecting prcccDNA and pCMV-Cre plasmids, to see whether the expression of HBV cccDNA was affected. The abundance of HBeAg in supernatant was measured by ELISA two days later. Compared to that in 19p13.11-ko Huh7 cells with siRNA-mock, the expression of HBeAg decreased to 88.06% (p<0.05) in 19p13.11-ko Huh7 cells with siRNA-YY1. Similarly, the expression of HBeAg decreased to 91.92% (p<0.05) when YY1 was knock down in 19p13.11-ko HepG2 cells (Figure S1D). The further reduction of HBV expression suggested that YY1 might have role(s) in regulating HBV replication other than mediating the combination between 19p13.11 enhancer and HBV cccDNA.
Figure 4. YY1 is the structural regulator of 19p13.11 enhancer and HBV cccDNA. A: Schematic diagram of enChIP technique. B: dCas9 protein (170kDa) – HBV DNA (130bp) – YY1 protein (65kDa) – 19p13.11 DNA (189bp) complex was detected by gRNA-HBV, and the abundance of YY1 protein and 19p13.11 DNA were both reduced when the binding site of YY1 was mutated in HBV (C) or the expression of YY1 was knock down (D) in Huh7 cells. The most obvious print in WB-YY1 was the heavy chain of antibody (55kDa).
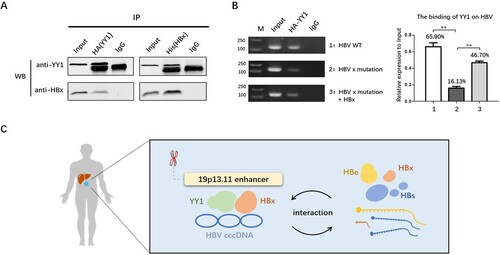
HBx could interact with YY1 and prompt the binding of YY1 to HBV genome
HBx has been reported to affect HBV cccDNA treancription by interacting with cellular factors. To explore whether HBx could interact with YY1, and particularly, the spatial proximity between HBV DNA and 19q13.11 enhancer element, pCMV-HA-YY1 and pCMV-His-HBx plasmids were co-transfected in Huh7 cells and Co-IP assay was conducted. The results showed that YY1 could bind to HBx protein whenever by anti-HA, or vice versa, by anti-His antibody. Moreover, when we transfected Huh7 cells with a prcccDNA plasmid containing a point mutation preventing HBx expression, the binding of YY1 on HBV cccDNA was decreased. And when the HBx protein was re-provided, the binding of YY1 on HBV cccDNA was partially recovered (B). These data suggested that HBx might play an important role on the binding ability of YY1 to HBV cccDNA.
Figure 5. HBx could interact with YY1 and promote the expression of YY1. A: co-IP experiments show that HBx protein (17kd) could be detected by anti-HA antibody and YY1 protein (65kd) could be detected by anti-His antibody. B: Chip-PCR experiments show the binding of YY1 on HBV cccDNA in Huh7 cells transfecting wild HBV plasmid, HBx-mutated plasmid with or without His-HBx plasmid (130bp). * P<0.05. ** P<0.01. C: Schematic diagram of spatial interaction between 19p13.11 enhancer and HBV cccDNA.
Discussion
Due to its long half-life and internal supplement mechanism of cccDNA, CHB is difficult to be cured. It has been accepted that, in addition to eradication, the permanent silence of cccDNA might be an effective and more realistic way for the cure of CHB. Therefore, it is important and worthwhile to explore the regulatory mechanism of cccDNA transcription and replication. More complicated, numerous studies had found that chromosomes of eukaryotic cells or yeast cells would present a highly complex three-dimensional structure, which may bring widely separated functional elements into close spatial proximity and create interactions among these elements located in the same or even different chromosome. Interestingly, HBV cccDNA could also be organized into viral-minichromosome similar with host chromosomes in structure and function, though the minichromosome is so small and the nucleosome spacing (repeat length) is 180 bp, instead of 200 bp repeat length for the chromatin of eukaryotic cells. Consequently, it is reasonable to speculate whether HBV cccDNA in minichromosome form has the same spatial interaction with the host chromosomes.
The 3C technique (chromosome conformation capture), via sequential formaldehyde cross linking, fragmentation and re-ligation of the cross-linked chromatin, allows identification of chromosomal elements interacted with each other in term of spatial proximity [Citation21]. Considering the difficulty to establish HBV infection cell models for continues passage, even by using of HepG2-NTCP and PHH under our conditions, in this study we took the advantage of the recombinant covalently closed circular DNA (rcccDNA) cell model for producing a cccDNA surrogate [Citation10–20]. By 3C technique, we did detect the spatial interaction between HBV minichromosome and 19p13.11 locus of host chromosome in Huh7 and HepG2 cells. Further analysis showed that the 19p13.11 locus is an active enhancer element and knockout of the ∼7 kb 19p13.11 enhancer region lead to reduced expression of HBV total RNA HBsAg and HBeAg in Huh7 and HepG2 cells (D). We also conducted pcDNA3.1-precore plasmid or pcDNA3.1-HBs plasmid transfection in wild type or 19p13.11-ko Huh7 and HepG2 cells respectively to confirm the specificity that only cccDNA can be recruited to 19p13.11, not a plasmid linked to a small part of the HBV genome. Alternatively, in future studies the 19p13.11 enhancer element can be knocked out from HepG2.2.15 cells where the HBV DNA is stably integrated into the host chromosome, which could test the specific interaction between 19p13.11 and HBV cccDNA once again. Thus, we concluded that 19p13.11 enhancer might play an important role in regulating the transcriptional activity of HBV cccDNA.
YY1 is a very known DNA binding factor involved in chromatin spatial interaction. As a transcript factor with high abundance in many different types of tissues and cells, YY1 could promote or repress the transcriptional activity of targeted sites via regulating histone acetylation or deacetylation [Citation27, Citation28]. Previous studies have found that YY1 is a structural regulator of enhancer-promoter chromatin loops [Citation16,Citation29], and the expression of YY1 was increased during HBV infection [Citation30]. Additionally, YY1 could prompt the formation of HBV cccDNA [Citation31]. However, whether YY1 protein involved in chromatin spatial interaction between HBV cccDNA and human genome has not been reported before. In this study, we demonstrated that YY1 protein mediated the spatial close of HBV cccDNA to 19p13.11 enhancer by a series of experiments, which was consistent to the results from 3C technique. Moreover, the binding site of YY1 in HBV genome is located at enhancer I region, which is quite conservative among different genotypes of HBV, showing the key role of YY1 on HBV cccDNA during its evolution progress. And correspondingly, when the expression of YY1 was knock down, the transcription activity of HBV cccDNA was also decreased. Thus, we demonstrated that YY1 could regulate the spatial distance of 19p13.11 enhancer or other host DNA elements to HBV cccDNA.
Though lacking of DNA binding capacity, HBV x protein is also involved in the activity of HBV cccDNA through interacting with other factors such as p300 and HDAC1 [Citation32]. In this study, we found that HBx could interact with YY1, and promote the binding of YY1 on HBV cccDNA. Since HBx also comes from cccDNA transcription, it seems that YY1 forms a positive feedback loop with HBV cccDNA to regulate HBV replication, suggesting the complexity and adaptability in the interaction between HBV and host (C).
However, the current work was based on human hepatoma cell lines, with rcccDNA (not true cccDNA) produced at very high (not physiological) level. It's well worth using liver biopsies or HepG2-NTCP cells infected with HBV, where true cccDNA is formed at low copy numbers per cell, to validate our results and look forward to more discoveries in future studies.
In summary, we demonstrated that the enhancer element in 19p13.11 region could up-regulate the transcriptional activity of HBV cccDNA through spatial interaction mediated by host YY1 protein and viral protein HBx, which will further refine the transcriptional regulation mechanism of HBV cccDNA.
Acknowledgments
This work was supported by the National S & T Major Project for Infectious Diseases (No. 2017ZX10201201), Beijing Municipal Natural Science Foundation (No. 7182080), the National Natural Science Foundation of China (No. 81672013 and 81974309), China Postdoctoral Science Foundation funded project (No. 2019M660363).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Wei XL, Luo HY, Li CF, et al. Hepatitis B virus infection is associated with younger median age at diagnosis and death in cancers. Int J Cancer. 2017;141(1):152–159.
- World Health Organization. Hepatitis B 2018. Available from: http://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
- Gane EJ. Future anti-HBV strategies. Liver Int 2017;37:40–44.
- McNaughton AL, Arienzo VD, Ansariet MA, et al. Insights from deep sequencing of the HBV genome-unique, tiny, and misunderstood. Gastroenterology. 2019;156(2):384–399.
- Wang J, Huang H, Liu Y, et al. HBV genome and life cycle. Adv Exp Med Biol. 2020;1179:17–37.
- Zhang Y, He S, Guo JJ, et al. Retinoid X receptor α-dependent HBV minichromosome Remodeling and viral replication. Ann Hepatol. 2017;16(4):501–509.
- Jain S, Chang TT, Chen S, et al. Comprehensive DNA methylation analysis of hepatitis B virus genome in infected liver tissues. Sci Rep. 2015;5:10478.
- Palumbo GA, Scisciani C, Pediconi N, et al. IL6 Inhibits HBV transcription by targeting the Epigenetic control of the nuclear cccDNA minichromosome. PLoS One. 2015;10(11):e0142599.
- Yuan Y, Zhao K, Yao Y, et al. HDAC11 restricts HBV replication through epigenetic repression of cccDNA transcription. Antiviral Res. 2019;172:104619.
- Du M, Bai L. 3D clustering of co-regulated genes and its effect on gene expression. Curr Genet. 2017;63(6):1017–1021.
- Zhang Y, Hyle J, Wright S, et al. A cis-element within the ARF locus mediates repression of p16 INK4A expression via long-range chromatin interactions. PNAS. 2019;116(52):26644–26652.
- Tam KT, Chan PK, Zhang W, et al. Identification of a novel distal regulatory element of the human neuroglobin gene by the chromosome conformation capture approach. Nucleic Acids Res. 2017;45(1):115–126.
- Shen C, Liu Y, Shi S, et al. Long-distance interaction of the integrated HPV fragment with MYC gene and 8q24.22 region upregulating the allele-specific MYC expression in HeLa cells. Int J Cancer. 2017;141(3):540–548.
- Tang Z, Luo OJ, Li X, et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015;163(7):1611–1627.
- Cattoglio C, Pustova I, Walther N, et al. Determining cellular CTCF and cohesin abundances to constrain 3D genome models. Elife. 2019;8:e40164.
- Beagan JA, Duong MT, Titus KR, et al. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017;27(7):1139–1152.
- Hensel KO, Cantner F, Bangert F, et al. Episomal HBV persistence within transcribed host nuclear chromatin compartments involves HBx. Epigenetics Chromatin. 2018;11(1):34.
- Moreau P, Cournac A, Palumbo GA, et al. Tridimensional infiltration of DNA viruses into the host genome shows preferential contact with active chromatin. Nat Commun. 2018;9(1):4268.
- Qi Z, Li G, Hu H, et al. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J Virol. 2014;88(14):8045–8056.
- Li G, Zhu Y, Shao D, et al. Recombinant covalently closed circular DNA of hepatitis B virus induces long-term viral persistence with chronic hepatitis in a mouse model. Hepatology. 2018;67(1):56–70.
- Dekker J, Rippe K, Dekker M, et al. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311.
- Jiao F, Shen C, Ning J, et al. HBV t1719g mutation reduced HBV replication through mutant Enh II and HBx protein in vitro. J Viral Hepat. 2019;26(6):710–717.
- Fujita T, Asano Y, Ohtsuka J, et al. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci Rep. 2013;3:3171.
- Jiang S, Yang Z, Li W, et al. Re-evaluation of the carcinogenic significance of hepatitis B virus integration in hepatocarcinogenesis. PLoS One. 2012;7(9):e40363.
- Raisner R, Kharbanda S, Jin L, et al. Enhancer activity requires CBP/P300 Bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24(7):1722–1729.
- Wang J, Xu ZW, Liu S, et al. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol. 2015;21(32):9554–9565.
- Khachigian LM. The Yin and Yang of YY1 in tumor growth and suppression. Int J Cancer. 2018;143(3):460–465.
- Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354(6350):241–245.
- Weintraub AS, Li CH, Zamudio AV, et al. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171(7):1573–1588.
- Shan X, Ren M, Chen K, et al. Regulation of the microRNA processor DGCR8 by hepatitis B virus proteins via the transcription factor YY1. Arch Virol. 2015;160(3):795–803.
- Hayashi Y, Kitamura Y, Nakanishi M, et al. The binding site of transcription factor YY1 is required for intramolecular recombination between terminally repeated sequences of linear replicative hepatitis B virus DNA. J Virol. 2000;74(20):9471–9478.
- Belloni L, Pollicino T, De Nicola F, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. PNAS. 2009;106(47):19975–19979.
