ABSTRACT
Candida albicans is a common fungal pathogen in humans that colonizes the skin and mucosal surfaces of the majority healthy individuals. How C. albicans disseminates into the bloodstream and causes life-threatening systemic infections in immunocompromised patients remains unclear. Plasminogen system activation can degrade a variety of structural proteins in vivo and is involved in several homeostatic processes. Here, for the first time, we characterized that C. albicans could capture and “subvert” host plasminogen to invade host epithelial cell surface barriers through cell-wall localized Eno1 protein. We found that the “subverted” plasminogen system plays an important role in development of invasive infection caused by C. albicans in mice. Base on this finding, we discovered a mouse monoclonal antibody (mAb) 12D9 targeting C. albicans Eno1, with high affinity to the 254FYKDGKYDL262 motif in α-helices 6, β-sheet 6 (H6S6) loop and direct blocking activity for C. albicans capture host plasminogen. mAb 12D9 could prevent C. albicans from invading human epithelial and endothelial cells, and displayed antifungal activity and synergistic effect with anidulafungin or fluconazole in proof-of-concept in vivo studies, suggesting that blocking the function of cell surface Eno1 was effective for controlling invasive infection caused by Candida spp. In summary, our study provides the evidence of C. albicans invading host by “subverting” plasminogen system, suggesting a potential novel treatment strategy for invasive fungal infections.
Introduction
Candida albicans is a common fungal microorganism that colonizes the oral, genital and gastrointestinal surfaces in most healthy individuals [Citation1,Citation2]. Colony maintenance requires a delicate balance between fungal proliferation and host immune recognition [Citation3]. In immunocompromised patients, C. albicans may disseminate into the bloodstream and lead to life-threatening systemic candidiasis [Citation4,Citation5]. However, anticandidal therapy is often of limited effectiveness in these patients, resulting in unacceptably high rates of mortality and morbidity. The associated mortality rate for systemic infection is reported to be greater than 30%, highlighting the potential critical impact of C. albicans on global health burden [Citation1,Citation6]. There is a justifiable need for more research to develop novel, more efficacious antifungal treatments.
In addition to host immunological defences, physical barriers also exist between different tissues to prevent dissemination of microorganisms [Citation7]. It takes several steps for C. albicans to establish invasive infection, including damage and invasion of the mucosal epithelium, vascular dissemination and seeding of yeast cells into the bloodstream, and target tissue invasion and colonization. The key for C. albicans to establish an invasive infection is damage and invasion of epithelial or endothelial cells [Citation8,Citation9]. Therefore, blocking this process might be an attractive approach for C. albicans caused invasive infections. Cell wall, the outermost cellular structure of C. albicans, is composed of cross-linked polysaccharides and glycoproteins, which are not only critical for the integrity and shape of the fungal cells as they grow and differentiate, but also a weapon for invading host [Citation8]. The exact mechanism of C. albicans cell wall damaging the host physical barriers has not been well characterized.
The physiological function of plasmin is integral to various homeostatic processes including blood coagulation, cell migration, and tissue and wound repair [Citation10]. The extracellular matrix (ECM) is the acellular protein component of animal tissues. It defines the basement membrane (BM) region and constitutes an anchoring platform for the epithelium, and is a part of the connective tissues that fill the interstitial spaces in the tissue parenchyma [Citation11,Citation12]. The ECM is basically composed of fibronectin (Fn), laminin (Ln), and collagen. Plasminogen is synthesized in the liver and released into the bloodstream as a zymogen with high concentrations (around 180 mg/mL) [Citation13]. The zymogen is converted into active plasmin protease by tissue-type plasminogen activator (tPA) and urokinase plasminogen activator (uPA) [Citation11]. Plasmin is a broad-specificity serine protease that degrades Fn, Ln, collagen, and a variety of other structural proteins. Plasmin can also activate other proteolytic enzymes, such as matrix metalloproteinases (MMPs), which act to degrade the tight junction components of microvascular endothelial cells [Citation13]. It has been reported that some invasive pathogens, such as Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenza, can associate with host plasminogen to their surface, and then converted plasminogen into active plasmin for degrading host ECM proteins and cell barriers [Citation13–16]. This mechanism is known to allow pathogens to migrate from periodontal tissue to various host organs via bloodstream [Citation11]. Furthermore, several fungal pathogens including C. albicans, A. fumigatus and C. neoformans are also known to be able to bind plasminogen [Citation17–19]. However, the molecular mechanism of fungal cell walls interacting with host plasminogen and the role of this interaction in establishment of invasive infections have not been elucidated.
Enolase is a dimeric enzyme that catalyses the penultimate step in glycolysis, interconverting 2-phosphoglycerate (2-PGA) and phosphoenolpyruvate (PEP) in cytoplasm [Citation20,Citation21]. Interestingly, localization of enolase to cell surface has been observed in several bacteria such as Borrelia burgdorferi, Staphylococcus aureus, Group A streptococci and Listeria monocytogenes [Citation22–25]. C. albicans cell surface located Eno1 has been identified as a moon-lighting protein with unrelated glycolytic enzyme function such as transglutaminase activity and major antigen in patients with candidiasis [Citation20,Citation21,Citation26]. Previous studies have demonstrated C. albicans Eno1 involved in colonization of mammalian intestinal epithelium and invasion of human brain microvascular endothelial cells in vitro [Citation27,Citation28]. In addition, Eno1 null mutant C. albicans exhibited avirulent in animal [Citation29]. However, the mechanism of Eno1 involved in the virulence of C. albicans remains unclear.
In the present study, we first characterized that C. albicans cell wall-localized Eno1 could capture and “subvert” host plasminogen for facilitating invasive infection. We further described a mouse monoclonal antibody (mAb) 12D9 targeting C. albicans Eno1, with a direct ability to block C. albicans capturing host plasminogen. Finally, we conducted proof-of-concept in vivo studies and showed that neutralization of cell surface Eno1 was effective for controlling C. albicans caused invasive infection.
Materials and methods
Ethics statement
All animal experiments were performed using procedures outlined by the “Regulations on the Administration of Laboratory Animals” approved by the State Council of the People's Republic of China. The animal experiment protocol has been verified and approved by the Animal Care and Use Committee of Tongji University (TJCAC-018-033).
Mice
Female C57BL/6 and BALB/c mice (6–8 weeks old) were obtained from Shanghai Laboratory Animal Center (SLAC) of the Chinese Academy of Sciences (Shanghai, China).
Reagents, antibodies and plasmids
IPTG (isopropyl-β-D-thiogalactopyranoside) and DTT were purchased from Sangon Biotech. Ni-nitrilotriacetic acid (Ni-NTA) was purchased from QIAGEN. Human plasminogen, ϵ-aminocaproic acid (ϵ-ACA), Creatinine Assay Kit, glass beads, urokinase-type plasminogen activator, anidulafungin, fluconazole, and DMEM medium were purchased from Sigma-Aldrich. Human Endothelial Serum Free Medium, HuMEC Basal Serum-Free Medium and Blood Urea Nitrogen Detection Kit were purchased from Thermo Fisher Scientific. Mouse nonspecific IgG2a was obtained from Invivogen. Cy3-labelled secondary antibody was purchased from Invitrogen. Chromogenic substrate D-Val-Leu-Lys-pNA·2HCl was purchased from Innovative Research. The LDH Cytotoxicity Assay Kit was obtained from Beyotime. PrimeScript TM RT Reagent Kit and the PrimeSTAR® Max DNA Polymerase were obtained from TaKaRa Bio. Rabbit anti-plasminogen antibody was obtained from Acris Antibodies. Anti-actin monoclonal antibody, and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG were obtained from Abcam. HRP-labelled goat anti-mouse antibody was purchased from Dingguochangsheng Biotechnology. pET-21a (+) was purchased from Novagen.
Candida spp. strains growth conditions
All strains were maintained on SDA agar plates (1% peptone, 4% dextrose, and 1.8% agar) and grown in YPD broth (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C. C. albicans SC5314 was kindly provided by Sanglard D (Centre Hospitalier Universitaire Vaudois). Clinical isolates C. albicans 0304103 and Y0109 were kindly provided by Dr. Jun Gu (Changhai Hospital, Shanghai, China). C. glabrata ATCC28226, C. krusei ATCC 6258, C. parapsilosis ATCC 34136 and C. tropical ATCC 20026 were obtained from ATCC.
For hyphal growth, exponentially growing C. albicans yeast cells were washed in PBS buffer and cultured in RPMI 1640 medium at 37°C for 3 h.
To obtain growth curve, exponentially growing C. albicans SC5314 were washed and resuspended in fresh YPD broth [optical density (OD)620 = 0.1] at 30°C with 200 rpm shaking, then the OD620 value was determined at the indicated time points.
Plasminogen-binding assay
Recombinant C. albicans Eno1 or other cell wall proteins (0.5 μg) were coated in 96-well plates (Nunc-Immuno) and blocked with PBS containing 5% bovine serum albumin (BSA). Then human plasminogen (0.5 μg per well) or negative control PBS buffer was added and incubated at 37°C for 1 h. Plasminogen binding was detected using a rabbit anti-plasminogen antibody (Acris Antibodies) and HRP-conjugated anti-rabbit IgG (Abcam). The reaction was quenched by 1 M H2SO4. Then the absorbance was measured at 450 nm using a multi-mode microplate reader.
For plasminogen binding to C. albicans assay, human plasminogen was labelled with allophycocyanin 650 according to the manufacturer’s instructions (Dojindo Molecular Technologies). Yeast (1 × 107 cells) or hyphal form (1 × 106 cells) of C. albicans were incubated in PBS containing allophycocyanin 650-labelled human plasminogen (5 μg/mL) at 37°C for 30 min. Then the binding of plasminogen with C. albicans was observed using a laser scanning confocal microscope (excitation wavelength, 650 nm; emission wavelength, 660 nm) (TCS SP5; Leica).
Plasminogen activity assay
The chromogenic substrate D-Val-Leu-Lys-pNA·2 HCl (Innovative Research) was dissolved in buffer (75 mM Tris HCl, 318 mM NaCl, pH 7.5) (0.25 mM), and treated with human plasminogen in the presence of C. albicans, non-albicans Candida spp. or other recombinant expressed C. albicans cell wall proteins. Urokinase-type plasminogen activator (0.1 g/mL) (Sigma-Aldrich) was used as positive control. The reaction mixture was incubated at 37°C for 24 h and then the absorbance was measured at 405 nm using a multi-mode microplate reader.
Host cell damage assay
Human Umbilical Vein Endothelial Cells (HUVECs) and Caco-2 intestinal epithelial cells were cultured with Human Endothelial Serum Free Medium and HuMEC Basal Serum-Free Medium in 96-well plate (1 × 104 cells per well) at 37°C with 5% CO2, respectively. Then C. albicans (2.5 × 103 cells per well) and Eno1 mAb or negative control mouse nonspecific IgG2a were added to the 96-well plates. After 12 h co-culture, cell supernatant was transferred for lactate dehydrogenase (LDH) assay using commercial kit (Beyotime). Maximal LDH release was determined by adding an LDH releasing agent (Beyotime) in parallel. Relative LDH release was measured as follows: LDH (%) = (OD490 indicated cells – OD490 control)/ (OD490 maximal LDH release – OD490 control).
Murine systemic candidiasis model
For the Candida spp. caused systemic infection in vivo, 6–8 weeks old C57BL/6 female mice were intravenously injected with 200 μl of PBS buffer containing indicated live Candida spp. cells [1 × 106 Colony Forming Unit (CFU) per mouse] for C. albicans, (C. tropicalis and C. parapsilosis; 1 × 107 CFU per mouse for C. glabrata; 5 × 105 CFU per mouse for C. krusei.). Then the survival was monitored for 30 days. And the kidneys and liver were removed 2 days post-infection and then homogenized in PBS buffer to determine fungal burden or fixed in 10% neutral formalin for haematoxylin and eosin (H&E) or periodic acid-schiffs (PAS) staining. Renal function tests were carried out using the Blood Urea Nitrogen Detection Kit (Thermo Fisher Scientific) and Creatinine Assay Kit (Sigma-Aldrich). To abolish plasminogen activity in vivo, mice were intraperitoneally injected with ϵ-ACA (30 mg/kg) twice daily for 5 days.
Western blot
C. albicans cells were collected and then resuspended in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM DTT, 0.5 mM PMSF, and 5 mg/mL of the protease inhibitors leupeptin, pepstatin, and antipain, pH 7.4). The cells were then lysed with glass beads (0.40 mm in diameter, Sigma-Aldrich) in a cell homogenizer (Braun, MSK). The insoluble fraction containing cell wall components was harvested by centrifuging at 8,000 g; the pellet was then washed with ice-cold water and boiled with extraction buffer (50 mM Tris-HCl, pH 7.5, 0.1 M EDTA, 2% SDS) to obtain cell wall proteins for immunoblot analysis. The cell wall proteins were subjected to SDS-PAGE, blotted with the Eno1 immune serum from mice (1:1000 dilution rate) and secondary antibodies, and then developed with the chemiluminescence method according to the manufacturer’s instructions (Millipore) using the ECL detection system (GE Healthcare). The densitometry of indicated blot was quantified using Image J software (National Institutes of Health, USA).
Quantitative real-time PCR
HUVECs or Caco-2 intestinal epithelial cells (2 × 107 cells) were challenged by exponentially growing C. albicans (MOI = 1) for 1 or and 3 h respectively. Then the total mRNA of C. albicans SC5314 was extracted using the PureLink™ RNA Mini Kit (Invitrogen) according to the manufacturer’s protocol. RT–PCR analysis was performed using the PrimeScript TM RT Reagent Kit (TaKaRa Bio). The individual gene-specific primers are listed in Table S1.
Expression and purification of C. albicans cell wall proteins
The gene encoding C. albicans Eno1 or its different domain, and other cell wall proteins were amplified from C. albicans SC5314 genomic DNA (the primers are listed in Table S2) and cloned into pET-21a (+) containing 6×His-tag. And then the plasmids were transformed into BL21 (DE3) pLysS cells for protein expression. The transformants were cultured overnight at 37°C and diluted 1:100 in fresh LB culture. When the medium OD600 was up to 0.6 at 37°C, IPTG was added at a final concentration of 0.1 mM and the transformants were grown at 16°C for 16 h. After that the transformants were lysed by sonication, and the target protein was purified by Ni-NTA (Qiagen).
Generation and selection of monoclonal antibodies against C. albicans Eno1
BALB/c mice (6–8 weeks old) were immunized with C. albicans Eno1 combined with complete Freund’s adjuvant or incomplete Freund’s adjuvant at multiple sites. Spleens from immunized mice were collected and underwent fusion with P3X63Ag8.653 myeloma cells to generate B-cell hybridomas [Citation30]. Hybridoma supernatants were screened for activity of binding with Eno1 using ELISA method and inhibiting C. albicans Eno1 binding to human plasminogen. The most active hybridomas were subjected to limiting dilution to obtain mAb. As a result, mAb 12D9 was selected for further study.
Enzyme-linked immunosorbent assay (ELISA)
Recombinant C. albicans or non-albicans Candida spp. Eno1, indicated domains or mutants of C. albicans Eno1 were coated in 96-wells plates (Thermo Scientific, 442404) (0.25 μg/well) overnight and blocked with PBS containing 5% BSA. The plates were then subjected to a series dilution of anti-Eno1 antibody and incubated overnight. After incubation with an HRP-labelled goat anti-mouse antibody (Dingguochangsheng Biotechnology) for 1 h, TMB substrate was added and incubated for 15 min incubation, and then 1 M H2SO4 were added to stop the reaction. Finally, the absorbance of reaction mixture was measured at 450 nm using a multi-mode microplate reader. 96-well microplate coated with Bovine Serum Albumin (BSA) was applied as parallel control in the analysis of plasminogen binding to recombinant C. albicans proteins. The test data of parallel control was deducted to eliminate the non-specific binding of plasminogen or antibodies to the microplate.
Surface plasmon resonance (SPR) analysis
Biacore T200 instrument (GE Healthcare) combined with a CM5 sensor chip (GE Healthcare) was used for SPR analysis. C. albicans Eno1 protein was immobilized in parallel-flow channel on a BIAcore™ CM5 sensor chip using the Amine Coupling Kit (GE Healthcare). Serial dilutions of mAb 12D9 were injected into the flow system. Experiments were conducted using PBS as running buffer, and the analyte was injected at a flow rate of 30 μl/min. The association time was 90 s and the dissociation time was 60 s. The affinity constants for binding were obtained using BIA evaluation software and a 1:1 Langmuir binding model.
Confocal laser scanning microscopy and flow cytometry analysis
Exponentially growing C. albicans SC5314 cells were incubated with mAb 12D9 or negative control mouse nonspecific IgG2a at 16°C overnight. Then the C. albicans SC5314 cells were washed with PBS and incubated with a Cy3-labelled secondary antibody (Invitrogen) at 30°C for 1 h. The stained C. albicans SC5314 cells were then mounted to microscope slides and analysed with a confocal laser scanning microscope (TCS SP5; Leica). For flow cytometry analysis, the stained cells were fixed with 1% formaldehyde overnight and analysed by flow cytometry (BD FACSVerse).
mAb opsono-phagocytic and killing assay
Thioglycollate-elicited peritoneal macrophages and neutrophils were isolated as previously described [Citation31]. For phagocytosis assay, exponentially growing C. albicans SC5314 cells were co-cultured with peritoneal macrophages (1 × 106 cells) (MOI = 0.4) in the presence of indicated concentrations of m Ab 12D9 at 37°C for 1 h. Then the unbound C. albicans SC5314 were removed by three washes of PBS buffer. Then the mixture was plated on SDA agar and incubated at 30°C for 48 h and the live C. albicans were counted.
For neutrophils killing assay, exponentially growing C. albicans SC5314 (1 × 104 cells) were incubated with indicated concentrations of mAb 12D9 at 30°C for 1 h. Then the C. albicans cells were collected and co-cultured with peritoneal neutrophils (2 × 106 cells) with MOI = 0.05 at 4°C for 1 h to make the cells settle, and then transferred to 37°C for another 1 h for killing test. During the incubation, control plates were placed at 4°C in parallel, and then the mixture were plated on SDA agar and incubated at 30°C for 48 h and the live C. albicans were counted.
C. albicans Eno1 modelling
C. albicans Eno1 structural model was built using the automated protein structure modelling server Swiss-Model (https://swissmodel.expasy.org/). BLAST and HHblits was performed for template search in the Swiss-Model template library. The model was built based on a targeted-template alignment using ProMod3. The geometry of the resulting model was regularized using a force field [Citation32–35].
Statistical analysis
At least three biological replicates were performed for all experiments unless otherwise indicated. Log-rank test was used for survival data analysis. For parametric data, the two-tailed Student’s t-test was used for analysis of two groups and one-way analysis of variance (ANOVA) was used for analysis of multiple groups. For nonparametric data, the nonparametric t-test or ANOVA was used. P value<0.05 was considered statistically significant.
Results
C. albicans activates host plasminogen to facilitate invasive infection
Although previous study reported C. albicans could bind human plasminogen, no direct evidence for a role in establishing invasive infection was found. We first investigated that whether C. albicans could activate the host plasminogen system to facilitate invasive infection. We initially labelled human plasminogen with allophycocyanin 650 and observed its binding to C. albicans using confocal laser scanning microscopy. C. albicans can grow in yeast and hyphal forms, and the hyphal form has an important role in causing disease by invading epithelial cells and causing tissue damage [Citation8]. The results revealed that plasminogen markedly bound to the surface of C. albicans SC5314 in both yeast and hyphae form, providing evidence that C. albicans may capture human plasminogen efficiently ((A) and Supplementary Figure S1). We further found C. albicans could convert human plasminogen to activate plasmin, determined by assaying its proteolytic activity on the chromogenic substrate D-Val-Leu-Lys-pNA·2 HCl, and this conversion effect could be abrogated by aminocaproic acid (ϵ-ACA, a known inhibitor of plasminogen activation) ((B,C)). Activated plasmin is a broad-specificity serine protease which could degrade several structural proteins. We found C. albicans could activate plasminogen to facilitate it to damage host endothelial cells, as determined by assaying LDH release after a 12-h co-culture of HUVECs with C. albicans (multiplicity of infection (MOI) = 0.1) in the presence of plasminogen ((D)). To further confirm whether host plasminogen activation in vivo facilitates C. albicans infection, the mice were treated with ϵ-ACA to abolish plasminogen activity. Although ϵ-ACA has no effect on the growth of C. albicans (Supplementary Figure S2), our data indicated that ϵ-ACA treatment significantly improved the survival of mice infected by C. albicans SC5314 ((E), 1 × 106 CFU per mouse). ϵ-ACA treatment also significantly reduced kidney fungal burdens of mice infected by C. albicans SC5314 ((F)). Therefore, the kidney functions of mice infected by C. albicans SC5314 were significantly improved after ϵ-ACA treatment, as indicated by the lower level of blood urea nitrogen (BUN) and creatinine (CRE) ((G,H)). The above results suggested that C. albicans could activate host plasminogen for facilitating invasive infection.
Figure 1. C. albicans activates host plasminogen to promote invasive infection. (A) Representative confocal microscope analysis of human plasminogen binding with C. albicans SC5314. Scale bar represents 10 μm. (B, C) Assay for yeast or hyphal form of C. albicans SC5314-induced plasminogen activation and the effect of plasminogen activation inhibitor ϵ-ACA (20 mM). (D) Plasminogen promoted the damage effects of C. albicans on HUVECs and the effect of ϵ-ACA (20 mM). HUVECs damage was determined by assaying LDH release after 12 h co-cultured with C. albicans SC5314 (MOI = 0.1). (E-F) C57BL/6 mice were treated ϵ-ACA (30 mg/kg) or vehicle control twice daily for 5 days after intravenously infection with C. albicans SC5314 (1 × 106 CFU per mouse, E-H). (E) Survival of the mice was monitored for 30 days. (F) The kidney fungal burden of the mice was determined at day 2 post-infection. (G, H) Blood urea nitrogen (BUN) and creatinine (CRE) levels in mice were determined at day 2 post-infection. UI, uninfected mice; PLG, Plasminogen. Data are represented as means ± SD from triplicates of one representative experiment of three. * P, < 0.05; ** P, < 0.01; *** P, < 0.001 [One-way ANOVA (B, C, D, G, H); Log-rank test (E); Nonparametric t-test (F)].
![Figure 1. C. albicans activates host plasminogen to promote invasive infection. (A) Representative confocal microscope analysis of human plasminogen binding with C. albicans SC5314. Scale bar represents 10 μm. (B, C) Assay for yeast or hyphal form of C. albicans SC5314-induced plasminogen activation and the effect of plasminogen activation inhibitor ϵ-ACA (20 mM). (D) Plasminogen promoted the damage effects of C. albicans on HUVECs and the effect of ϵ-ACA (20 mM). HUVECs damage was determined by assaying LDH release after 12 h co-cultured with C. albicans SC5314 (MOI = 0.1). (E-F) C57BL/6 mice were treated ϵ-ACA (30 mg/kg) or vehicle control twice daily for 5 days after intravenously infection with C. albicans SC5314 (1 × 106 CFU per mouse, E-H). (E) Survival of the mice was monitored for 30 days. (F) The kidney fungal burden of the mice was determined at day 2 post-infection. (G, H) Blood urea nitrogen (BUN) and creatinine (CRE) levels in mice were determined at day 2 post-infection. UI, uninfected mice; PLG, Plasminogen. Data are represented as means ± SD from triplicates of one representative experiment of three. * P, < 0.05; ** P, < 0.01; *** P, < 0.001 [One-way ANOVA (B, C, D, G, H); Log-rank test (E); Nonparametric t-test (F)].](/cms/asset/f536fb80-c34c-4da0-a078-9c894c350608/temi_a_1840927_f0001_oc.jpg)
Eno1 plays a central role in C. albicans capturing and activating of host plasminogen
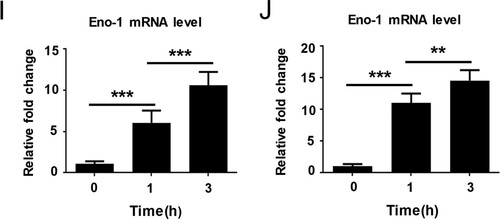
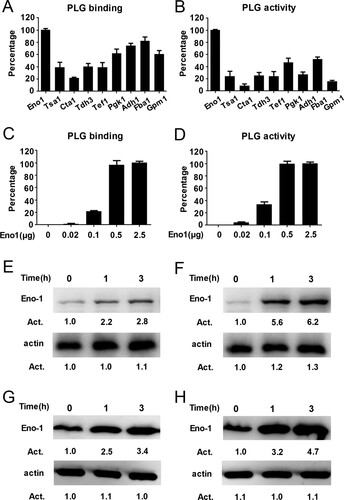
Previous studies have identified nine possible plasminogen-binding proteins on the surface of C. albicans, including Eno1, Tsa1, Cta1, Tdh3, Tef1, Pgk1, Adh1, Fba1, and Gpm1 [Citation17,Citation21]. We used a bacterial expression system to produce high purity of the above possible plasminogen-binding proteins and further compared their plasminogen binding and activating ability (Supplementary Figure S3(A), (B)). We found that C. albicans Eno1 displayed the strongest plasminogen binding and activating ability among these proteins ((A,B)). Further data indicated that the C. albicans Eno1 bound to and activated plasminogen in a dose-dependent manner ((C,D)). To investigate the role of Eno1 during C. albicans invasion of the host cell, we assayed C. albicans Eno1 expression during invasion of HUVECs and Caco-2 intestinal epithelial cells. The results indicated that the expression of Eno1 were significantly increased at protein level (cell wall and cytoplasmic located) ((E,F) and (G,H)) and mRNA level ((I,J)) ((E,G,I) for C. albicans SC5314 invading HUVECs; (F,H,J) for C. albicans SC5314 invading Caco-2 intestinal epithelial cells) with the infecting time increased. These findings suggested that C. albicans Eno1 plays a central role in capturing and activating host plasminogen to promote invasion of host cells.
Figure 2. Eno1 plays a central role in C. albicans activating human plasminogen. (A) ELISA assays for recombinant C. albicans cell wall proteins binding to human plasminogen. Recombinant C. albicans Eno1 binding to human plasminogen was regarded as 100 percent. (B) Assays for the ability of recombinant C. albicans cell wall proteins to activate human plasminogen. Recombinant C. albicans Eno1 activating human plasminogen was regarded as 100 percent. (C) ELISA assays for recombinant C. albicans Eno1 binding to human plasminogen. Recombinant C. albicans Eno1 (2.5 μg) binding to human plasminogen was regarded as 100 percent. (D) Assays for recombinant C. albicans Eno1-induced activation of human plasminogen. Recombinant C. albicans Eno1 (2.5 μg) activating plasminogen was regarded as 100 percent. (E-J) Eno1 expression at protein level and mRNA level during C. albicans SC5314 invading human endothelial or epithelial cells in vitro. HUVECs (E, G, I) and Caco-2 intestinal epithelial cells (F, H, J) were co-cultured with C. albicans SC5314 for the indicated time (MOI = 1). C. albicans cell wall-localized (E, F) or cytoplasmic (G, H) Eno1 protein were determined by western blot and Eno1 mRNA expression levels were determined by Q-PCR (I, J). PLG, Plasminogen. Data shown in (A-D), (I) and (J) are means ± SD of triplicates from one representative experiment of three. The immunoblotting analysis shown in (E-H) are representative of three independent experiments. ** P, < 0.01 *** P, < 0.001 (One-way ANOVA).
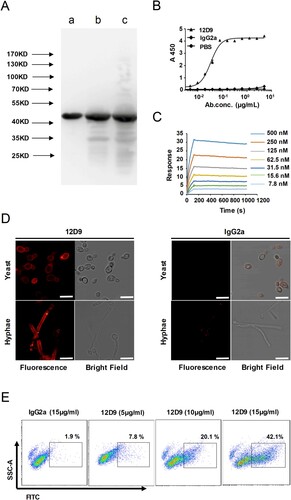
mAb 12D9 binds dose-dependently, specifically to C. albicans Eno1 with high affinity, and blocks C. albicans from capturing plasminogen
Anti-Eno1 mAbs were generated in BALB/c mice which were immunized with recombinant C. albicans Eno1. Hybridomas were generated from the splenocytes of the immunized mice and P3X63Ag8.653 myeloma cells. Initially, >200 hybridoma supernatants were detected to contain IgGs which could bind with recombinant C. albicans Eno1 by ELISA. The hybridoma supernatants with significant affinity to C. albicans Eno1 were screened for ability to inhibit C. albicans Eno1 binding and activating of human plasminogen; whereby the pool of hybridomas expressing functional IgGs was reduced to 11. mAb 12D9 was selected from the 11 anti-Eno1 mAb based on its blocking the recombinant C. albicans Eno1 function of binding to plasminogen (Supplementary Figure S4). The results of western blot indicated that mAb 12D9 could specifically bind with Eno1 protein from C. albicans cytoplasm and cell wall ((A)). The kinetics of mAb 12D9 binding to recombinant C. albicans or non-albicans Candida spp. Eno1 was determined using ELISA (EC50 = 0.03–0.07 μg/ml) ((B) and Supplementary Figure S5) and SPR analysis ((C)). The SPR analysis indicated mAb 12D9 had a high affinity with recombinant C. albicans Eno1 [association rate constant (ka), 4.34 × 104 (1/M/s); dissociation rate constant (kd), 7.44 × 10−5 (1/s)]. And the estimated KD (equilibrium dissociation constant) for mAb 12D9 binding to recombinant C. albicans Eno1 was 1.711 nM. A therapeutic delivery of mAb must be able to bind its target cells. Therefore, mAb 12D9 binding to C. albicans was assessed by confocal microscopy and flow cytometry. Confocal microscopy revealed that mAb 12D9 bound to the yeast and hyphae surface of C. albicans markedly ((D)) and flow cytometry analysis indicated this binding was mAb 12D9 dose-dependent ((E)). Taken together, our data indicate that mAb 12D9 displays a high affinity with recombinant C. albicans Eno1 and could prevent C. albicans from capturing plasminogen.
Figure 3. mAb 12D9 targeting C. albicans Eno1 displayed high affinity with recombinant Eno1 and C. albicans cell. (A)Western blot analysis to verify the specificity of mAb 12D9. Recombinant C. albicans Eno1 (Band a), cell wall extracts (Band b), and cytoplasmic extracts (Band c) from C. albicans SC5314 were fracted by SDS-PAGE and then subjected to immunoblotting analysis with mAb 12D9. The western bolt analysis are representative of three independent experiments. (B) ELISA assay for mAb 12D9 binding to recombinant C. albicans Eno1. (C) SPR analysis for interaction between the mAb 12D9 and recombinant C. albicans Eno1. Data shown in (B) and (C) come from one representative experiment of three. (D) Representative confocal microscope analysis for mAb 12D9 binding to C. albicans SC5314. Scale bar represents 10 μm. (E) Representative images analysed by flow cytometry for detection of mAb 12D9 binding to C. albicans. Data shown in (D, E) are representative images of three experiments.
Furthermore, we found that mAb 12D9 could not only block recombinant C. albicans Eno1 binding and activating of plasminogen ((A,B)), but also block the plasminogen activating ability of C. albicans SC5314, clinical isolate C. albicans Y0109 and non-albicans Candida spp. ((C), Supplementary Figures S6 and S7).
Figure 4. mAb 12D9 could block C. albicans capturing and activating human plasminogen. The assay for C. albicans Eno1 binding to (A) and activating (B) human plasminogen in the presence of the indicated concentration of mAb 12D9. Recombinant C. albicans Eno1 binding to and activating plasminogen in the absence of mAb 12D9 were regarded as 100 percent. (C) The assays for C. albicans SC5314 activating human plasminogen in the presence of mAb 12D9 (10μg). PLG, Plasminogen. Bla, Blank. ** P, < 0.01[One-way ANOVA (A, C)].
![Figure 4. mAb 12D9 could block C. albicans capturing and activating human plasminogen. The assay for C. albicans Eno1 binding to (A) and activating (B) human plasminogen in the presence of the indicated concentration of mAb 12D9. Recombinant C. albicans Eno1 binding to and activating plasminogen in the absence of mAb 12D9 were regarded as 100 percent. (C) The assays for C. albicans SC5314 activating human plasminogen in the presence of mAb 12D9 (10μg). PLG, Plasminogen. Bla, Blank. ** P, < 0.01[One-way ANOVA (A, C)].](/cms/asset/0123aefa-bee3-4cdb-bebe-334388405deb/temi_a_1840927_f0004_ob.jpg)
mAb 12D9 blocks C. albicans capturing plasminogen in mice to reduce invasive infection
Damage of epithelial or endothelial cells is the key step of C. albicans developing invasive infection. Therefore, to investigate whether mAb 12D9 could inhibit C. albicans induced infection, we first tested the effect of mAb 12D9 on C. albicans damage of HUVECs and Caco-2 intestinal epithelial cells. In the presence of plasminogen, we found that mAb 12D9 could significantly reduce C. albicans medicated damage (diminished LDH release) on HUVECs and Caco-2 cells ((A,B) and Supplementary Figure S8). While, the inhibitory effect was not observed in the absence of plasminogen with a dose-dependent manner ((A,B)). Neutrophils and macrophages are the primary leukocytes responsible for phagocytosis and killing of invasive fungal cells. Antibody binding to pathogens can induce host innate immune cell-mediated phagocytosis and killing. mAb 12D9 mediated fungal opsono-phagocytosis and killing were measured by co-culture of C. albicans and macrophages or neutrophils in the presence of mAb 12D9. We found that mAb 12D9 exhibited dose-dependent opsono-phagocytosis and killing activity when macrophages and neutrophils were challenged by C. albicans SC5314 ((C,D)).
Figure 5. mAb 12D9 blocks C. albicans developing invasive infection. (A, B) HUVECs (A) and Caco-2 intestinal epithelial cells (B) damage induced by C. albicans in the presence of plasminogen and mAb 12D9 (10 μg/mL) was determined by assaying LDH release. Relative levels of LDH release from HUVECs or Caco-2 intestinal epithelial cells were measured after 6 h of co-culture with C. albicans (MOI = 0.1). (C) Phagocytosis of C. albicans by thioglycollate-elicited peritoneal macrophages. C. albicans SC5314 was co-cultured with peritoneal macrophages and the indicated concentration of mAb 12D9 at 37°C for 1 h (MOI = 0.4). The suspension was then plated on SDA agar for 48 h, after which C. albicans colonies were counted and the phagocytosis percent was calculated. (D) Killing of C. albicans by thioglycollate-elicited peritoneal neutrophil. C. albicans SC5314 cells were co-cultured with peritoneal neutrophils and the indicated concentration of mAb 12D9 at 37°C for 1 h. The suspension was then plated on SDA agar for 48 h, after which C. albicans colonies were counted and the killing percent was calculated. Data in (A-D) are represented as means ± SD from triplicates of one representative experiment of three. (E-G) C57BL/6 mice were infected with 1 × 106 CFU of C. albicans SC5314 and treated with mAb 12D9 (30 mg/kg) and/or anidulafungin (AN) (0.1 mg/kg) via the lateral tail vein. (E) Survival of mice was monitored for 30 days (n = 10 per group). (F) Quantification of the fungal burden in the kidneys of mice (n = 6 per group) at day 2 post-infection. (G) Representative H&E (for determining inflammatory cell influx and the extent of tissue necrosis) and PAS (for C. albicans) staining of kidneys from infected mice with the indicated treatment at day 2 post-infection. Arrows indicate inflammatory cells influx and tissue necrosis (H&E staining) and C. albicans filaments in the tissues (PAS staining). Magnification 200×. Data in (G-J) are representative of three independent experiments. *, P < 0.05; ***, P < 0.001[One-way ANOVA (A-D); Log-rank test (E); Nonparametric One-way ANOVA (F)].
![Figure 5. mAb 12D9 blocks C. albicans developing invasive infection. (A, B) HUVECs (A) and Caco-2 intestinal epithelial cells (B) damage induced by C. albicans in the presence of plasminogen and mAb 12D9 (10 μg/mL) was determined by assaying LDH release. Relative levels of LDH release from HUVECs or Caco-2 intestinal epithelial cells were measured after 6 h of co-culture with C. albicans (MOI = 0.1). (C) Phagocytosis of C. albicans by thioglycollate-elicited peritoneal macrophages. C. albicans SC5314 was co-cultured with peritoneal macrophages and the indicated concentration of mAb 12D9 at 37°C for 1 h (MOI = 0.4). The suspension was then plated on SDA agar for 48 h, after which C. albicans colonies were counted and the phagocytosis percent was calculated. (D) Killing of C. albicans by thioglycollate-elicited peritoneal neutrophil. C. albicans SC5314 cells were co-cultured with peritoneal neutrophils and the indicated concentration of mAb 12D9 at 37°C for 1 h. The suspension was then plated on SDA agar for 48 h, after which C. albicans colonies were counted and the killing percent was calculated. Data in (A-D) are represented as means ± SD from triplicates of one representative experiment of three. (E-G) C57BL/6 mice were infected with 1 × 106 CFU of C. albicans SC5314 and treated with mAb 12D9 (30 mg/kg) and/or anidulafungin (AN) (0.1 mg/kg) via the lateral tail vein. (E) Survival of mice was monitored for 30 days (n = 10 per group). (F) Quantification of the fungal burden in the kidneys of mice (n = 6 per group) at day 2 post-infection. (G) Representative H&E (for determining inflammatory cell influx and the extent of tissue necrosis) and PAS (for C. albicans) staining of kidneys from infected mice with the indicated treatment at day 2 post-infection. Arrows indicate inflammatory cells influx and tissue necrosis (H&E staining) and C. albicans filaments in the tissues (PAS staining). Magnification 200×. Data in (G-J) are representative of three independent experiments. *, P < 0.05; ***, P < 0.001[One-way ANOVA (A-D); Log-rank test (E); Nonparametric One-way ANOVA (F)].](/cms/asset/604ac82a-c66e-478f-942c-47a634af98ef/temi_a_1840927_f0005_oc.jpg)
To determine the antifungal infection effect of mAb 12D9 in vivo, mice were infected via intravenously injection of C. albicans SC5314 [1 × 106 Colony-Forming Units (CFU) per mouse] and treated with mAb 12D9 (30 mg/kg). The data showed that mAb 12D9 could significantly improve the survival of mice ((E)) and reduced fungal burden in the kidneys ((F)). Additionally, mAb 12D9 also displayed significantly synergistic antifungal activity with a sub-effective dose of anidulafungin ((F)) or fluconazole (Supplementary Figure S9(A–C)). H&E staining revealed that inflammatory influx was significantly diminished and PAS staining showed less hyphae in the kidneys of mice treated with mAb 12D9 ((G)) (Supplementary Figure S9(D)). Furthermore, mAb 12D9 could also reduce kidney fungal burden of mice infected by clinical isolates C. albicans Y0109 and 0304103 (Supplementary Figure S10).
Although C. albicans remains the predominant fungal species that causes systemic and mucosal infections, non-albicans Candida spp. are also regarded as an important cause of serious candidemia [Citation36]. Therefore, we also determined the effect of mAb 12D9 on non-albicans Candida spp. caused infections. We found that treatment with mAb 12D9 could significantly reduce the kidney fungal burdens in mice with candidemia caused by Candida parapsilosis, Candida tropicalis, Candida glabrata or Candida krusei (Supplementary Figure S11).
Our above results indicated that mAb 12D9 is a promising potential therapeutic antibody for both C. albicans and non-albicans Candida spp. caused invasive infections.
Maximal activity of mAb 12D9 against C. albicans infection depends on host plasminogen activation
To gain further insight into the mechanism of how mAb 12D9 against C. albicans infection, we examined whether the antifungal activity of mAb 12D9 is dependent on host plasminogen activation. We found that mAb 12D9 could inhibit C. albicans induced damage to HUVECs (determined via LDH release) in the presence of plasminogen, while the inhibit effect was abolished when the activation of plasminogen was inhibited by ϵ-ACA ((A)). We further abolished plasminogen in mice by ϵ-ACA treatment (30 mg/kg). As a result, mAb 12D9 failed to improve survival and reduce fungal burden in the kidneys and liver of C. albicans infected mice with ϵ-ACA treatment ((B–D)). In addition, no significant improvement of kidneys function (BUN and CRE) was detected in the experimental mice ((E,F)). The above results suggested that the effect of mAb 12D9 against fungal infection depends on host plasminogen activation.
Figure 6. Maximal activity of mAb 12D9 against C. albicans infection depends on host plasminogen. (A) HUVECs damage was determined by assaying LDH release. Relative levels of LDH release from HUVECs were measured after 6 h of co-culture with C. albicans (MOI = 0.1) in the presence of indicated plasminogen, mAb 12D9 (4μg/mL) and ϵ-ACA (20 mM). (B-F) C57BL/6 mice were treated with ϵ-ACA (30 mg/kg) twice daily for 5 days to abolish plasminogen system after intravenously infection with C. albicans SC5314 (1 × 106 CFU). (B) Survival of mice with the indicated mAb 12D9 (30 mg/kg) and/or anidulafungin (AN) (0.1 mg/kg) treatment was monitored for 30 days (n = 10 per group). (C, D) Quantification of fungal burden in kidneys (C) and liver (D) of mice treated with mAb 12D9 (30 mg/kg) and/or anidulafungin (0.1 mg/kg) (n = 6 per group) at day 2 post-infection. (E, F) Blood urea nitrogen (BUN) (E) and creatinine (CRE) (F) in mice were determined at day 2 post-infection. UI, uninfected mice. Data in (A-F) are representative of three independent experiments. **, P < 0.01; ***, P < 0.001; [Nonparametric One-way ANOVA (A, C-F); Log-rank test (B)].
![Figure 6. Maximal activity of mAb 12D9 against C. albicans infection depends on host plasminogen. (A) HUVECs damage was determined by assaying LDH release. Relative levels of LDH release from HUVECs were measured after 6 h of co-culture with C. albicans (MOI = 0.1) in the presence of indicated plasminogen, mAb 12D9 (4μg/mL) and ϵ-ACA (20 mM). (B-F) C57BL/6 mice were treated with ϵ-ACA (30 mg/kg) twice daily for 5 days to abolish plasminogen system after intravenously infection with C. albicans SC5314 (1 × 106 CFU). (B) Survival of mice with the indicated mAb 12D9 (30 mg/kg) and/or anidulafungin (AN) (0.1 mg/kg) treatment was monitored for 30 days (n = 10 per group). (C, D) Quantification of fungal burden in kidneys (C) and liver (D) of mice treated with mAb 12D9 (30 mg/kg) and/or anidulafungin (0.1 mg/kg) (n = 6 per group) at day 2 post-infection. (E, F) Blood urea nitrogen (BUN) (E) and creatinine (CRE) (F) in mice were determined at day 2 post-infection. UI, uninfected mice. Data in (A-F) are representative of three independent experiments. **, P < 0.01; ***, P < 0.001; [Nonparametric One-way ANOVA (A, C-F); Log-rank test (B)].](/cms/asset/b3b36d78-9964-457b-8c0d-0e195e830a1c/temi_a_1840927_f0006_oc.jpg)
mAb 12D9 binds to a motif in the helix 6 sheet 6 loop of C. albicans Eno1 to prevent it capturing and activating host plasminogen
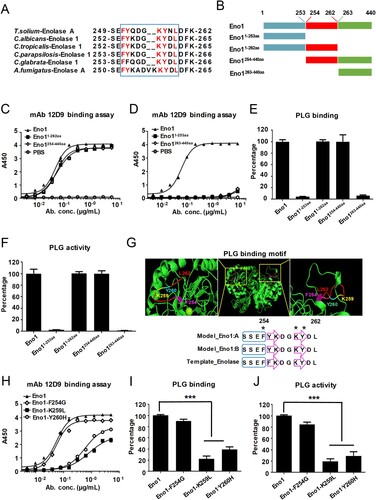
The 251FYQDGKYNL259 sequence in the helix 6 sheet 6 (H6S6) loop of T. solium EnoA has been identified as the putative plasminogen binding motif [Citation37]. We aligned the putative plasminogen-binding loop of T. solium EnoA with enolase of fungal species including C. albicans, C. tropicalis, C. parapsilosis, C. glabrata and A. fumigatus. High similarity of plasminogen-binding motif was identified among these enolase sequences ((A)). Specifically, the 254FYKDGKYDL262 motif in the H6S6 loop of C. albicans Eno1 shared high sequence identity with the putative plasminogen-binding loop motif of T. solium EnoA ((A)). In order to map the plasminogen-binding sites of C. albicans Eno1 and the antigen epitope of mAb 12D9 binding to, four domains of Eno1 were recombinant expressed: domain 1-253aa, domain 1-262aa, domain 254-440aa and domain 263-440aa ((B) and Supplementary Figure S3(C)). We subsequently investigated the affinity of mAb 12D9 for each of the four domains of C. albicans Eno1. The data indicated that mAb 12D9 had a similar affinity for C. albicans Eno11-262aa and Eno1254-440aa, as well as C. albicans Eno1. However, mAb 12D9 exhibited low affinity for C. albicans Eno11-253aa and Eno1263-440aa ((C,D)). Furthermore, C. albicans Eno11-262aa and Eno1263-440 could bind and activate human plasminogen, while C. albicans Eno11-253aa and Eno1263-440aa could not bind and activate human plasminogen ((E,F)). The above data suggested mAb 12D9 targets to motif 254FYKDGKYDL262 in the H6S6 loop of C. albicans Eno1 to block it binding to and activating plasminogen. Three-dimensional structural model showed that residues F254, K259, and Y260 plays critical role for maintain the structure of motif 254FYKDGKYDL262 ((G)). Based on this, we recombinant expressed three mutants of C. albicans Eno1(F254G, K259L and Y260H). We found that mAb 12D9 displayed decreased affinity with the Eno1-K259L and Eno1-Y260H mutants but had a similar affinity for the Eno1-F254G mutant compared to C. albicans Eno1 ((H)). Furthermore, the Eno1-K259L and Eno1-Y260H mutants exhibited significantly lower plasminogen binding and plasminogen activating abilities, while similar levels of plasminogen binding and activation of the Eno1-F254G mutant were detected compared to C. albicans Eno1 ((I,J)). Our results revealed that mAb 12D9 binds to a motif (254FYKDGKYDL262) with key residues of K259 and Y260 in the H6S6 loop of C. albicans Eno1 to prevent it capturing and activating host plasminogen.
Figure 7. mAb 12D9 binds motif 254FYKDGKYDL262 in H6S6 loop of C. albicans Eno1 to prevent it activating plasminogen. (A) Alignment of the H6S6 loop amino acid sequences of T. solium enolase A with enolase of fungal species including C. albicans, C. tropicalis, C. parapsilosis, C. glabrata and A. fumigatus. (B) Schematic diagram of different domains of recombinant C. albicans Eno1. (C, D) ELISA assay for mAb 12D9 binding to C. albicans Eno1 and its different domains (Eno1254-440aa and Eno11-262aa, C; Eno1263-440aa and Eno11-253aa, D). (E) Plasminogen-binding assay with different domains of recombinant C. albicans Eno1. (F) Plasminogen activation induced by different domains of recombinant C. albicans Eno1. (G) Structural model of C. albicans Eno1 (homo-dimer) generation using Swiss-Model. Putative Eno1-plasminogen interaction motif was marked in red, and the key plasminogen-binding residues (F254G, amaranth; K259L, yellow; Y260H, cyan) were calculated by Swiss-Model and marked with “*”. A blue box stands for α-helices and a purple arrow represents a β-strand. (H) ELISA assay for mAb 12D9 binding to recombinant C. albicans Eno1 and mutations (F254G, K259L and Y260H). (I) Plasminogen-binding assay for recombinant C. albicans Eno1 and mutations (F254G, K259L and Y260H). (J) Plasminogen activation induced by recombinant C. albicans Eno1 and mutations (F254G, K259L and Y260H). Recombinant C. albicans Eno1 binding to and activating plasminogen in the absence of mAb 12D9 were regarded as 100 percent. PLG, Plasminogen. Data in (C-F and H-J) are representative of three independent experiments. *** P, < 0.001 (One-way ANOVA).
Discussion
Physical barriers play an important role in host controlling dissemination of pathogens. Invasion and damaging host physical barriers are the premise of C. albicans progressing from colonization to systemic infection. Previous study suggested that Eno1 is involved in C.albicans colonization and invasion of host cells barriers in vitro [Citation27,Citation28]. Our findings demonstrated that the C. albicans cell wall-localized Eno1 can capture and manipulate host plasminogen system to damage mucosal epithelium and vascular endothelial cells. We further discovered that mAb 12D9 which targets to Eno1 could prevent host plasminogen being captured by C. albicans and was effective for controlling candidemia in vivo.
The mammalian plasminogen-plasmin system plays a central role in fibrinolysis, ECM degradation, and cell migration [Citation13]. Previous studies have suggested that pathogens such as B. burgdorferi and S. aureus can activate host plasminogen system to degrade the ECM of mucosal epithelium for promoting invasive infections [Citation18,Citation38]. Here, we found that C. albicans can efficiently capture and activate of human plasminogen system ((A–C) and Supplementary Figure S1). Since C. albicans colonizes human mucosal surfaces, its ability to bind to and activate plasminogen may be an important pathogenic mechanism. We found that plasminogen binding makes C. albicans more virulent in the HUVECs-C. albicans interaction model in vitro ((D)).
Previous work has demonstrated that ϵ-ACA can effectively inhibit activity of plasminogen [Citation39]. Therefore, we abolished plasminogen-plasmin system using ϵ-ACA in mice to explore the role of plasminogen activation in C. albicans caused systemic infection in vivo. Our results demonstrated that ϵ-ACA could lessen C. albicans systemic infection in mice and highlight an important role of plasminogen activation in C. albicans establishing systemic infection ((E–H)).
Enolase catalyses the conversion of 2-PGA to PEPE and its activity is critical for both glycolysis and gluconeogenesis across virtually all taxa. However, it has been reported that enolase is also cell wall-localized as a multifunctional protein with non-glycolytic functions in several pathogens [Citation21–23]. α-Enolase has been identified as a primary plasminogen receptor on the surface of streptococcal groups [Citation21,Citation40–42]. Eno1 was responsible for transglutaminase activity on cell wall of C. albicans and acts as the major antigen in patients with candidiasis. Both passive and active vaccinations with C. albicans Eno1 display protective effect against disseminated candidiasis [Citation43,Citation44]. The role of enolase in pathogenicity of a variety of fungal species, including Aspergillus nidulans, Aspergillus fumigatus, C. albicans and Paracoccidioides brasiliensis has been reported [Citation21,Citation45,Citation46]. C. albicans Eno1 null mutant exhibits abnormal hyphal formation, attenuated virulence and is more susceptible to antifungal agents such as amphotericin B and azoles [Citation29]. In this study, we found that Eno1 displayed the strongest plasminogen-binding and activation activity in the nine possible plasminogen receptors of C. albicans ((A–D)). Our finding demonstrated that Eno1 expression was significantly increased during C. albicans invading host cell ((E–J)). Hence, we propose that Eno1 plays the most important role in pathogenicity of C. albicans by activating host plasminogen. However, Tef1 and Adh1 also displayed plasminogen binding and activating ability which should not be neglected ((A,B)), and it will be investigated in our future study.
Neutralization is the most direct mechanism by which antibodies counteract pathogens. The Fc domain of antibodies can also recruit innate immune cells to engage in effector functions such as opsono-phagocytosis and cytotoxicity [Citation47]. Our study identified and characterized mAb 12D9 with high affinity to C. albicans and non-albicans Candida spp. Eno1 ((A–C) and Supplementary Figure S5). We found that mAb 12D9 could markedly bind to C. albicans and block it capturing and activating plasminogen ((D,E), and Supplementary Figure S6). Since C. albicans-mediated activation of host plasminogen could promote invasive infections, we investigated the anti-infective effect of mAb 12D9 using an in vitro host cell-C. albicans interaction model. We found that mAb 12D9 could attenuate the damage of C. albicans on endothelial or epithelial cells ((A–D)), as well as promote recruitment of macrophages and neutrophils to target C. albicans via its Fc domain ((E,F)). As mAb 12D9 exerts both Eno1 blocking and opsono-phagocytic effects, we further confirmed these in vitro findings using a systemic infection mouse model. The experiments indicated that mAb 12D9 monotherapy or combination therapy with anidulafungin or fluconazole displayed significant antifungal activity in mice ((E–G)) (Supplementary Figure S9). Since enolase is a highly conserved protein across Candida spp. It is reasonable that mAb 12D9 also has antifungal effects in mice with non-albicans Candida spp. caused invasive infection (Supplementary Figure S11). Together, our data suggested that mAb 12D9 may be a potential agent for invasive Candida spp. infections.
Blasting peptide sequence in Candida Genome Database (http://www.candidagenome.org) demonstrated that Eno1 is the only protein in C. albicans containing FYKDGKYDL sequence. Alignment analysis revealed that the 254FYKDGKYDL262 motif in C. albicans Eno1 shared high sequence identity with a putative plasminogen-binding motif of T. solium Enolase A (251FYQDGKYNL259) ((A,B)). By testing the affinity of mAb 12D9 and various recombinant C. albicans Eno1 domains, we confirmed that 254FYKDGKYDL262 motif in C. albicans Eno1 was the antigen epitope of mAb 12D9 ((B–D)). Our data further indicated that the motif 254FYKDGKYDL262 is also required for C. albicans Eno1 binding and activation of plasminogen ((E,F)). To date, the crystal structure of C. albicans Eno1 has not been reported. It is classified by the CATH database as a member of the enolase superfamily with the code 3.20.20.120, which indicates that the C. albicans Eno1 proteins consists mainly of α-helices and β-strands and has a TIM-type α-β-barrel. Swiss-Model analysis revealed that the motif 254FYKDGKYDL262 is located in the H6S6 loop of C. albicans Eno1, and suggested that residue F254 in α-helix 6 and residues K259 and Y260 in β-sheet 6 were important for the secondary structure of this domain ((G)). The analysis interaction of Eno1 F254G, K259L and Y260H mutants with human plasminogen confirmed that residues K259 and Y260 are key for Eno1 induced human plasminogen activation ((G)). Affinity analysis indicated that Eno1 K259 and Y260 were required for mAb 12D9 binding ((H–J)).
Invasive candidiasis is an important health-care-associated fungal infection and is widely recognized as a major cause of morbidity and mortality in the health-care environment despite antifungal therapy. Our present study identified that C. albicans have the ability to capture and “subvert” host plasminogen system to facilitate tissue invasion, and indicated that cell wall-localized Eno1 plays a central role in this process. Base on this finding, we further characterized the mAb 12D9 with high affinity to the 254FYKDGKYDL262 motif in H6S6 loop of C. albicans Eno1. mAb 12D9 showed its ability to prevent host plasminogen being captured and “subverted” by C. albicans, providing a potential novel treatment strategy for invasive fungal infections.
Table_S2.docx
Download ()Table_S1.docx
Download ()Figure_S11.docx
Download ()Figure_S10.docx
Download ()Figure_S9.docx
Download ()Figure_S8.docx
Download ()Figure_S7.docx
Download ()Figure_S6.docx
Download ()Figure_S5.docx
Download ()Figure_S4.docx
Download ()Figure_S3.docx
Download ()Figure_S2.docx
Download ()Figure_S1.docx
Download ()Acknowledgements
We would like to thank Dr Sanglard D (Centre Hospitalier Universitaire Vaudois) for kindly providing C. albicans SC5314, and Dr Jun Gu (Changhai Hospital, Shanghai, China) for kindly providing the C. albicans isolates 0304103 and Y0109. This study was supported by the National Natural Science Foundation of China (81671989, 81471924, 81903670, 81601745), Scientific research projects of the Shanghai Science and Technology Committee (18411951300), Shanghai Youth Medical Talents Training Program (Laboratory Medicine) [2018]02, China Postdoctoral Science Foundation (2019M651590), and the Key Projects of the 12th Five Year of the Chinese Army (BWS12J027). Mao-Mao An, Si-Min Chen, Hui Shen, and Zui Zou conceptualized the study. Si-Min Chen, Hui Shen. Shi-Yu Guo, Wei-Tong Hou, Xi-Ran Qiu, Yu Zhang, Jun-Li Song, and Xin-Yu Hu performed the experiments. Mao-Mao An, Si-Min Chen, Hui Shen, and Yuan-Ying Jiang supervised the study and wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kullberg BJ, Arendrup MC. Invasive candidiasis. N Engl J Med. 2015 Oct 8;373(15):1445–1456.
- Stop neglecting fungi. Nat Microbiol. 2017 Jul 25;2:17120.
- Netea MG, Joosten LA, van der Meer JW, et al. Immune defence against Candida fungal infections. Nat Rev Immunol. 2015 Oct;15(10):630–642.
- LeibundGut-Landmann S, Wuthrich M, Hohl TM. Immunity to fungi. Curr Opin Immunol. 2012 Aug;24(4):449–458.
- Erwig LP, Gow NA. Interactions of fungal pathogens with phagocytes. Nat Rev Microbiol. 2016 Mar;14(3):163–176.
- McCarty TP, Pappas PG. Invasive candidiasis. Infect Dis Clin North Am. 2016 Mar;30(1):103–124.
- Lemichez E, Lecuit M, Nassif X, et al. Breaking the wall: targeting of the endothelium by pathogenic bacteria. Nat Rev Microbiol. 2010 Feb;8(2):93–104.
- Gow NA, van de Veerdonk FL, Brown AJ, et al. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 2012 Feb;10(2):112–122.
- Yano J, Lilly E, Barousse M, et al. Epithelial cell-derived S100 calcium-binding proteins as key mediators in the hallmark acute neutrophil response during Candida vaginitis. Infect Immun. 2010 Dec;78(12):5126–5137.
- Chapin JC, Hajjar KA. Fibrinolysis and the control of blood coagulation. Blood Rev. 2015 Jan;29(1):17–24.
- Ayon-Nunez DA, Fragoso G, Bobes RJ, et al. Plasminogen-binding proteins as an evasion mechanism of the host's innate immunity in infectious diseases. Biosci Rep. 2018 Oct 31;38(5):BSR20180705.
- Peetermans M, Vanassche T, Liesenborghs L, et al. Bacterial pathogens activate plasminogen to breach tissue barriers and escape from innate immunity. Crit Rev Microbiol. 2016 Nov;42(6):866–882.
- Lahteenmaki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 2005 Feb;13(2):79–85.
- Sun H, Ringdahl U, Homeister JW, et al. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004 Aug 27;305(5688):1283–1286.
- Agarwal V, Kuchipudi A, Fulde M, et al. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J Biol Chem. 2013 Mar 8;288(10):6849–6863.
- Barthel D, Singh B, Riesbeck K, et al. Haemophilus influenzae uses the surface protein E to acquire human plasminogen and to evade innate immunity. J Immunol. 2012 Jan 1;188(1):379–385.
- Crowe JD, Sievwright IK, Auld GC, et al. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol Microbiol. 2003 Mar;47(6):1637–1651.
- Zaas AK, Liao G, Chien JW, et al. Plasminogen alleles influence susceptibility to invasive aspergillosis. PLoS Genet. 2008 Jun 20;4(6):e1000101.
- Stie J, Fox D. Blood-brain barrier invasion by Cryptococcus neoformans is enhanced by functional interactions with plasmin. Microbiology (Reading). 2012 Jan;158(Pt 1):240–258.
- Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, et al. alpha-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol. 2012;2012:156795.
- Funk J, Schaarschmidt B, Slesiona S, et al. The glycolytic enzyme enolase represents a plasminogen-binding protein on the surface of a wide variety of medically important fungal species. Int J Med Microbiol. 2016 Jan;306(1):59–68.
- Nogueira SV, Smith AA, Qin JH, et al. A surface enolase participates in Borrelia burgdorferi-plasminogen interaction and contributes to pathogen survival within feeding ticks. Infect Immun. 2012 Jan;80(1):82–90.
- Molkanen T, Tyynela J, Helin J, et al. Enhanced activation of bound plasminogen on Staphylococcus aureus by staphylokinase. FEBS Lett. 2002 Apr 24;517(1-3):72–78.
- Pancholi V. Fischetti VA. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998 Jun 5;273(23):14503–14515.
- Schaumburg J, Diekmann O, Hagendorff P, et al. The cell wall subproteome of Listeria monocytogenes. Proteomics. 2004 Oct;4(10):2991–3006.
- Reyna-Beltran E, Iranzo M, Calderon-Gonzalez KG, et al. The Candida albicans ENO1 gene encodes a transglutaminase involved in growth, cell division, morphogenesis, and osmotic protection. J Biol Chem. 2018 Mar 23;293(12):4304–4323.
- Jong AY, Chen SH, Stins MF, et al. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J Med Microbiol. 2003 Aug;52(Pt 8):615–622.
- Silva RC, Padovan AC, Pimenta DC, et al. Extracellular enolase of Candida albicans is involved in colonization of mammalian intestinal epithelium. Front Cell Infect Microbiol. 2014;4:66.
- Ko HC, Hsiao TY, Chen CT, et al. Candida albicans ENO1 null mutants exhibit altered drug susceptibility, hyphal formation, and virulence. J Microbiol. 2013 Jun;51(3):345–351.
- Tkaczyk C, Hua L, Varkey R, et al. Identification of anti-alpha toxin monoclonal antibodies that reduce the severity of Staphylococcus aureus dermonecrosis and exhibit a correlation between affinity and potency. Clin Vaccine Immunol. 2012 Mar;19(3):377–385.
- Zhang SQ, Zou Z, Shen H, et al. Mnn10 maintains pathogenicity in Candida albicans by extending alpha-1,6-mannose backbone to evade host dectin-1 mediated antifungal immunity. PLoS Pathog. 2016 May;12(5):e1005617.
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018 Jul 2;46(W1):W296–W303.
- Bienert S, Waterhouse A, de Beer TA, et al. The SWISS-MODEL repository – new features and functionality. Nucleic Acids Res. 2017 Jan 4;45(D1):D313–D319.
- Hu JL, Hua YJ, Chen Y, et al. Structural analysis of tumor-related single amino acid mutations in human MxA protein. Chin J Cancer. 2015 Sep 28;34(12):583–593.
- Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014 Jul;42(Web Server issue):W252–W258.
- Quindos G. Epidemiology of candidaemia and invasive candidiasis. A changing face. Rev Iberoam Micol. 2014 Jan-Mar;31(1):42–48.
- Ayon-Nunez DA, Fragoso G, Espitia C, et al. Identification and characterization of Taenia solium enolase as a plasminogen-binding protein. Acta Trop. 2018 Jun;182:69–79.
- Stie J, Bruni G, Fox D. Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion. PloS one. 2009 Jun 3;4(6):e5780.
- Berri F, Rimmelzwaan GF, Hanss M, et al. Plasminogen controls inflammation and pathogenesis of influenza virus infections via fibrinolysis. PLoS Pathog. 2013 Mar;9(3):e1003229.
- Derbise A, Song YP, Parikh S, et al. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect Immun. 2004 Jan;72(1):94–105.
- Pal-Bhowmick I, Vora HK, Jarori GK. Sub-cellular localization and post-translational modifications of the Plasmodium yoelii enolase suggest moonlighting functions. Malar J. 2007 Apr 16;6:45.
- Bergmann S, Wild D, Diekmann O, et al. Identification of a novel plasmin(ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Mol Microbiol. 2003;49(2):411–423.
- Li W, Hu X, Zhang X, et al. Immunisation with the glycolytic enzyme enolase confers effective protection against Candida albicans infection in mice. Vaccine. 2011 Jul 26;29(33):5526–5533.
- Xin H, Dziadek S, Bundle DR, et al. Synthetic glycopeptide vaccines combining beta-mannan and peptide epitopes induce protection against candidiasis. Proc Natl Acad Sci U S A. 2008 Sep 9;105(36):13526–13531.
- Marcos CM, de Fatima da Silva J, de Oliveira HC, et al. Surface-expressed enolase contributes to the adhesion of Paracoccidioides brasiliensis to host cells. FEMS Yeast Res. 2012 Aug;12(5):557–570.
- Yadav RK, Shukla PK. A novel monoclonal antibody against enolase antigen of Aspergillus fumigatus protects experimental aspergillosis in mice. FEMS Microbiol Lett. 2019 Feb 1;366(3):fnz015.
- Lu LL, Suscovich TJ, Fortune SM, et al. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol. 2018 Jan;18(1):46–61.