ABSTRACT
Shiga toxin (Stx)-producing Escherichia coli (STEC) can cause a wide range of symptoms from asymptomatic carriage, mild diarrhea to bloody diarrhea (BD) and hemolytic uremic syndrome (HUS). Intimin, encoded by the eae gene, also plays a critical role in STEC pathogenesis. Herein, we investigated the prevalence and genetic diversity of eae among clinical STEC isolates from patients with diarrhea, BD, HUS as well as from asymptomatic STEC-positive individuals in Sweden with whole-genome sequencing. We found that 173 out of 239 (72.4%) of clinical STEC strains were eae positive. Six eae subtypes (ϵ1, γ1, β3, θ, ζ and ρ) were identified eae and its subtype γ1 were significantly overrepresented in O157:H7 strains isolated from BD and HUS patients. ϵ1 was associated with O121:H19 and O103:H2 strains, and β3 to O26:H11 strains. The combination of eae subtype γ1 and stx subtype (stx2 or stx1+stx2) is more likely to cause severe disease, suggesting the possibility of using eae genotypes in risk assessment of STEC infection. In summary, this study demonstrated a high prevalence of eae in clinical STEC strains and considerable genetic diversity of eae in STEC strains in Sweden from 1994 through 2018, and revealed association between eae subtypes and disease severity.
Introduction
Shiga toxin (Stx)-producing Escherichia coli (STEC), is an enteric foodborne pathogen that can be asymptomatic or cause mild diarrhea, bloody diarrhea (BD) or even hemolytic uremic syndrome (HUS) in infected humans [Citation1,Citation2]. HUS is the leading cause of acute renal failure in children with high morbidity and mortality [Citation3]. Serotype O157:H7, associated with HUS and severe clinical outcomes, is the most predominant and virulent serotype among more than 400 serotypes that have been identified [Citation2,Citation4]. Nevertheless, since the early 2010s, non-O157 pathogenic serogroups, such as O26, O103 and O104, have been widely reported from HUS patients [Citation5–7]. Ruminants, especially cattle, are the most important reservoir of STEC [Citation8]. Direct contact with animals and their environment, consumption of undercooked beef, unpasteurized milk, other animal-derived products, contaminated water and vegetables are the main sources of human infections [Citation9].
Pathogenicity of STEC in human is largely dependent on Stx, which is considered as the most important virulence factor. Stx, which is encoded by stx genes located on lambdoid prophages, has two types, Stx1 and Stx2, where Stx2 shows much stronger correlation with severe symptoms [Citation10,Citation11]. The duration of stx shedding is a main cause of secondary person-to-person (fecal-oral) transmission, the longer duration poses a high transmission risk [Citation6]. Besides Stx, intimate adherence of STEC to the intestinal epithelium is also an important process in the STEC pathogenesis, it can cause attaching and effacing (A/E) lesions, which is a hallmark of STEC pathogenesis [Citation12,Citation13]. Intimin, encoded by the eae gene located in the locus of enterocyte effacement (LEE) pathogenicity island [Citation14], plays a determinant role in the formation of A/E lesions by inducing the effacement of microvilli and forming of actin pedestals [Citation15,Citation16]. Also, intimin cooperates with its translocated intimin receptor-Tir, to trigger host signalling events and actin nucleation, thus inducing lesion formation [Citation17]. Moreover, STEC injects a series of effector proteins into host cells through a type III secretion system (T3SS), which is encoded by LEE pathogenicity island, to play its pathogenic role [Citation18].
The full length of the eae gene is about 2,800 base pairs (bp). eae has several subtypes, owning to its heterogeneous 3’ regions, which encoded protein that has been identified to be the intimin cell-binding domain (Int280a) [Citation19]. There are at least 19 groups of eae subtypes, i.e. α, β, γ, ϵ, ξ, z, η, θ, τ, ι, κ, λ, μ, ν, υ, ο, π, ρ and σ, that have been defined so far [Citation20]. It has been suggested that intimin alleles are responsible for different host specificity and tissue tropism [Citation21]. Roger et al. showed that eae subtype γ1 appeared to be the most frequent among O157:H7 and O145:H28/H25/H- strains [Citation22,Citation23]. A previous study has investigated the genetic diversity of intimin in atypical Enteropathogenic Escherichia coli (EPEC), where intimin β1 was suggested to be the most frequent subtype among atypical EPEC strains from diarrheal patients [Citation24]. However, the molecular characteristics of eae-positive STEC strains, especially clinical strains, have rarely been described. Moreover, the relationship between eae subtypes and clinical symptoms, as well as duration of stx shedding remains to be addressed.
The aim of this study is therefore to investigate the eae subtypes and polymorphisms among clinical STEC strains isolated from STEC-positive individuals present with varying symptoms in Sweden, and to assess the association of eae subtypes with disease severity.
Materials and Methods
Ethics statement
The study was approved by the regional ethics committees in Gothenburg (2015/335-15) and Stockholm (2020-02338), Sweden, respectively.
Strain collection and clinical information
A total of 239 STEC strains were isolated from STEC-infected individuals from 1994 through 2018 in Sweden. Clinical data of STEC patients were collected through reviewing medical records as well as routine praxis used for the STEC surveillance performed in Sweden. The duration of bacterial shedding was determined as the time period from the first stx-PCR-positive sample to the first negative sample, and clinical symptoms were classified into HUS, bloody diarrhea (BD) and non-bloody stool (NBS) [Citation25].
Whole-genome sequencing, assembly and annotation
Bacterial DNA was extracted and whole genomes were sequenced by Illumina HiSeq X platform at SciLifeLab (Stockholm, Sweden) and Ion Torrent S5 XL platform (Thermo Fisher Scientific, Waltham, Massachusetts, US) at The Public Health Agency of Sweden as previously described [Citation26]. The Illumina sequencing reads were de novo assembled with SKESA (version 2.3.0) [Citation27]. The Ion Torrent sequencing reads were de novo assembled with SPAdes (version 3.12.0) in “careful mode” [Citation28]. The genome assemblies were annotated with Prokka (version 1.14.6) [Citation29]. The assemblies of all strains in this study were deposited in GenBank with accession numbers and metadata shown in Table S1.
Serotyping and stx subtyping
Serotype was determined by comparing assemblies to the SerotypeFinder database (DTU, Denmark) (http://www.genomicepidemiology.org/) using BLAST+ v2.2.30 [Citation30]. The stx subtypes were determined by ABRicae version 0.8.10 (https://github.com/tseemann/abricate). An in-house stx subtyping database was created with ABRicate by including representative nucleotide sequences of all identified stx1 and stx2 subtypes. The assemblies were then used to search against the stx subtyping database. Multilocus sequence typing (MLST) was conducted in silico using the on-line tool provided by the Warwick E. coli MLST scheme website (https://enterobase.warwick.ac.uk/species/ecoli/allele_st_search). Sequences types (STs) were determined based on the seven housekeeping allelic genes (adk, fumC, gyrB, icdF, mdh, purA, and recA) profile.
Eae subtyping and polymorphism analysis
The complete sequences of the eae gene were extracted from the genome sequences according to the genome annotation, and then aligned with reference sequences of all described eae subtypes downloaded from GeneBank. The genetic distances of eae subtypes were computed using the Maximum Composite Likelihood method by MEGA 7.0 software, and a Neighbor-Joining tree was generated with 1,000 bootstrap resamplings. As earlier described [Citation20], a 95% nucleotide sequence identity cut-off value was used to characterize an innovative eae subtype. eae genotypes (GTs) based on eae sequence polymorphism was used to determine the diversity within each eae subtype.
Comparison of clinical eae-positive STEC strains with strains from other sources
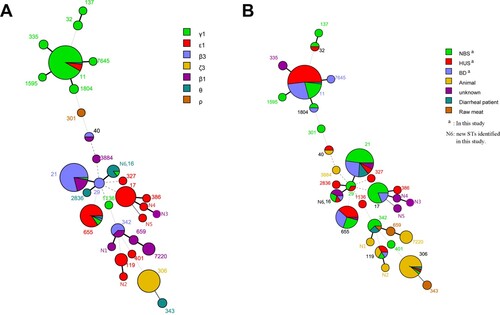
To assess the relationship of eae-positive clinical STEC strains in this study and strains from other sources, the MLST allelic profiles of eae-positive strains isolated from animals, meat and humans reported in a recent survey [Citation20] were used for comparison. A minimum spanning tree was generated with BioNumerics software version 7.6 (Applied Maths, Belgium).
Statistical analyses
Fisher's exact test was used to analyze the association between eae subtypes and bacterial features or clinical outcomes, the statistical significance was determined by Statistica12 (StatSoft, Inc. Tibco), p-value <0.05 was considered statistically significant.
Results
Prevalence of eae in clinical STEC strains
Among 239 clinical STEC strains, eae was present in 173 (72.4%) strains, including 56 HUS-associated strains, and 117 non-HUS strains (44 from patients with BD and 73 from individuals with NBS). All 65 O157:H7 strains and 108 (62.1%) non-O157 strains carried eae (p<0.0001). eae was overrepresented in strains from children (73.08%, p=0.011). The presence of eae was significantly associated with BD, HUS, and O157:H7 (). However, no association was observed between the presence of eae and the duration of bacterial shedding.
Table 1. Prevalence of eae gene in 239 STEC strains isolated from STEC-positive individuals.Table Footnotea
Diversity and subtype of eae in correlation with clinical outcomes
Six eae subtypes, namely epsilon1 (ϵ1), gamma1 (γ1), beta3 (β3), theta (θ), zeta3 (ζ3) and rho (ρ), were assigned in 173 eae-positive STEC strains. γ1 was present in 39.3% of all strains, being the most predominant subtype, followed by ϵ1 (30.6%) and β3 (24.9%). GTs were analyzed to determine the diversity within each eae subtype. Twenty-nine unique eae sequences were obtained, among which, ϵ1 subtype had ten GTs (GT1-GT10), followed by β3 (GT1-GT8), γ1 (GT1-GT5), θ (GT1-GT4), ζ3 (GT1) and ρ (GT1) (). Isolates with the same eae subtype clustered together with the corresponding references (). Notably, γ1 was statistically overrepresented in strains from BD and HUS patients, while β3 was related to non-HUS strains (). No association was found between eae subtypes and the duration of bacterial shedding or age of patients (Table S2).
Figure 1. Phylogenetic relationships of 29 different eae sequences identified in this study and 30 eae subtypes reference sequences based on Neighbor-Joining method. The corresponding eae subtype (number of strains), strain name, serotype (number of strains), and stx subtype (number of strains) are shown. The eae subtypes/genotypes in this study are indicated in bold and different colors. Scale bar indicates genetic distance.
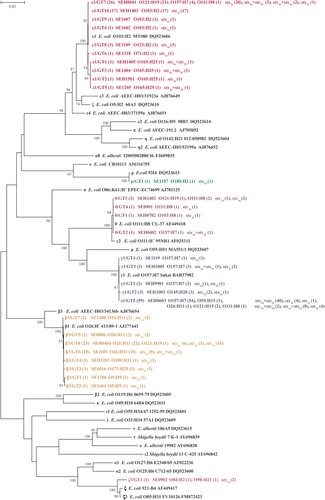
Table 2. Association between eae subtypes and clinical symptoms or bacterial variables.Table Footnotea
Eae subtypes and genotypes correlated with serotypes
In total, 173 eae-positive strains were typed into seventeen O serogroups and nine H types, which were assigned into 17 serotypes, O157:H7 (n=65, 37.57%) was the most predominant serotype. O157:H7 strains harbored eae-γ1 (60, 92.31%), ϵ1 (4, 6.15%) and θ (1, 1.54%). γ1 was found to be statistically associated with O157:H7, while ϵ1 and β3 were related to non-O157 serotypes (). Among non-O157 serotypes, we found that ϵ1 was associated with O121:H19 and O103:H2 serotypes, β3 was associated with O26:H11. The only two strains with eae subtype ζ3 were assigned to serotypes O84:H2 and O98:H21. Only O180:H2 strain carried eae subtype ρ (Table S3). All O121:H19 strains carried eae with one exception. ϵ1/GT7 was found to be linked to O121:H19 (p<0.0001).
Association of eae and its subtypes with stx subtypes
Overall, six stx subtypes and combinations were identified in 173 eae-positive STEC strains, namely, stx1a, stx2a+stx2c, stx2a, stx2c, stx1a+stx2a, and stx1a+stx2c, among which stx2 (57.2%) was more prevalent than stx1 (35.3%). We found that the presence of stx1+eae was significantly more prevalent in non-HUS STEC strains compared with HUS associated STEC strains. Notably, the presence of stx2+eae was significantly linked to HUS associated STEC strains, and stx1+stx2+eae was linked to BD-related strains ().
Table 3. Association between presence of stx + eae and clinical symptoms.
stx1a was the most predominant stx subtype, with 35.3% of strains carrying this subtype. stx1a+ stx2c only existed in strains carrying eae-γ1. eae-ρ subtype was only found in strains harboring stx2a. Notably, ϵ1 was associated with stx2a, γ1 was related to stx2a+stx2c, and stx1a+stx2c, β3 was linked to stx1a (Table S4).
Comparison of clinical eae-positive STEC strains with strains from other sources
Twenty-two sequence types (STs) were found in the 173 eae-positive STEC isolates (Table S1). ST11 was the most common sequence type, all 61 O157:H7 strains belonged to ST11. A minimum spanning tree was generated using 22 STs from this study and 18 STs from other sources reported previously [Citation20]. Interestingly, isolates from the same source showed tendency to cluster closely. For instance, isolates from humans, independent on patients with BD, HUS, or individuals with NBS, clustered closely, while isolates from animals and raw meat showed closer relatedness. Notably, when grouped with eae subtypes, the majority strains belonging to the most predominant eae subtype γ1 were grouped closely with a few exceptions. Similarly, strains with the same other eae subtypes were more likely to cluster closely ().
Discussion
STEC strains harboring eae are suggested to be more pathogenic with a higher risk of developing HUS [Citation31]. Little is known regarding the features and polymorphism of eae gene in STEC strains derived from patients as well as their association with disease severity. Here, we performed molecular characterization of eae-positive STEC strains from patients with a variety of symptoms as well as asymptomatic carriers. We found that 72.4% of clinical STEC strains were eae positive, out of which 37.6% were O157 strains, and 62.4% were non-O157 strains. All clinical O157 strains were eae positive, while 62.1% of non-O157 strains carried eae, which is much higher than that of reported in a recent study where only 9.5% of non-O157 strains carried eae [Citation20]. We found that 99.3% of HUS associated STEC strains possessed eae, which was significantly higher than eae prevalence in non-HUS STEC strains (65.4%). Additionally, eae positive rate in strains isolated from patients with BD (86.3%) was higher than that of individuals with NBS (57.0%). It’s well-recognized that O157 is the primary cause of HUS [Citation32–34]. We found that eae was significantly more prevalent in O157 strains, which may partially explain severe clinical outcomes of O157 strains.
The eae sequences in 173 STEC strains were classified into six subtypes, namely ϵ1, γ1, β3, θ, ζ3 and ρ. eae-γ1 and ϵ1 were the most common subtypes in this study. The prevalence of eae subtypes varies among studies. In a previous study, β1 and ζ3 were the most prevalent eae subtypes among STEC strains from different sources including diarrhea patients, raw beef and mutton, cattle, and yak [Citation20]. eae-γ1 and β1 were reported to be the most widespread subtypes in STEC strains isolated from patients in Germany [Citation35]. eae alleles examined in STEC strains isolated from ruminant animals also showed great genetic diversity. β and ζ were the most common eae subtypes in strains isolated from sheep, while β and θ were more prevalent in strains from cattle [Citation36]. ϵ1 and γ1 were the most frequent eae subtypes among STEC strains isolated from healthy cattle [Citation23]. The reason possibly lies in different sample sources and geographic distribution. It has been demonstrated that eae subtype β, ϵ, γ1, and θ are linked to more virulent strains [Citation37]. Here, we found that γ1 was associated with severe clinical symptoms such as BD and HUS, highlighting the clinical significance of eae subtype γ1. However, the underlying mechanisms how different eae subtypes modulate the pathogenicity remains to be elucidated.
A diverse range of serotypes were observed among eae-positive STEC isolates. An earlier study showed association between serotypes and eae subtypes: O157 and O145 strains tended to harbor γ1, O103 and O121 harbored ϵ, O26 carried β, while O111 possessed θ and β [Citation37]. In the present study, we observed a similar pattern. γ1 was significantly overrepresented in O157:H7 strains, also in line with a previous report of STEC strains derived from humans in Switzerland [Citation22]. Similarly, ϵ1 was found to be prevalent in O121:H19 and O103:H2 strains. β3 was predominant in O26:H11 strains.
Notably, ϵ1 was found in four O157:H7 strains, which has been rarely reported before. In Germany, two O157:H16 strains isolated from diarrheal children carried eae-ϵ [Citation38]. Another study reported O157:H16 strains isolated from water and meat also harbored eae-ϵ [Citation39]. More data is needed to characterize the O157:H7 strains carrying eae-ϵ subtype.
The coexistence of stx and eae, especially stx2, are more likely to enhance virulence and increase the severity of clinical outcomes in humans than those carrying stx1 alone [Citation40–42]. Consistently, we found that the presence of stx2+eae in STEC strains is strongly associated with HUS. Interestingly, the presence of stx1+stx2+eae was linked to BD, while stx1+eae was associated with NBS, supporting the evidence that the presence of stx2, rather than stx1, together with eae was associated with severe disease. Besides the finding that eae subtype γ1 was associated with HUS and O157:H7, γ1 was also found to be associated with stx2a+stx2c and stx1a+stx2c, these are high virulent stx subtypes, which could also contribute to the severity of clinical symptoms.
Longer duration of stx shedding poses higher risk for the transmission of STEC strains from person to person, STEC-infected patients below 15 years old are usually associated with longer shedding duration [Citation6,Citation43]. Consistently, we found that children had a longer shedding duration than adults (unpublished data). Several genes were reported to be associated with prolonged duration of shedding [Citation25]. In this study, we found that the presence of eae was associated with children. However, the presence of eae and subtypes has no association with the duration of stx shedding. As the information of age and the duration of shedding for some individuals is missing, further research is needed to understand the role of eae in children with longer duration of shedding.
In conclusion, here we describe the prevalence and genetic diversity of eae genes in clinical STEC isolates from Sweden from 1994 through 2018. Our results show that the majority of the clinical STEC isolates carry eae genes, which demonstrate highly genetic diversity. We found associations between eae subtypes and certain serotypes. Furthermore, eae subtype γ1 is associated with strains causing severe symptoms. However, no correlation was observed between the presence of eae gene/subtypes and duration of bacterial shedding. Our study proposes that the coexistence of eae subtype γ1 and stx2 or stx1+stx2, could be used as a risk predictors for severe symptoms of STEC infections.
Table_S4.xlsx
Download ()Table_S3.xlsx
Download ()Table_S2.xlsx
Download ()Table_S1.xlsx
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kakoullis L, Papachristodoulou E, Chra P, et al. Shiga toxin-induced haemolytic uraemic syndrome and the role of antibiotics: a global overview. J Infect. 2019;79(2):75–94.
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. The Lancet. 2005;365(9464):1073–1086.
- Wong CS, Jelacic S, Habeeb RL, et al. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections [Journal article; Research Support, U.S. Gov't, P.H.S.]. N Engl J Med. 2000 2000-06-29;342(26):1930–1936.
- Lynn RM, O'Brien SJ, Taylor CM, et al. Childhood hemolytic uremic syndrome, United Kingdom and Ireland. Emerging Infect. Dis.. 2005 2005-01-01;11(4):590–596.
- Delannoy S, Mariani-Kurkdjian P, Bonacorsi S, et al. Characteristics of Emerging human-pathogenic Escherichia coli O26:H11 strains isolated in France between 2010 and 2013 and carrying thestx2d gene only. J Clin Microbiol. 2015;53(2):486–492.
- Vonberg RP, Höhle M, Aepfelbacher M, et al. Duration of fecal shedding of Shiga toxin–producing Escherichia coli O104:H4 in patients infected During the 2011 Outbreak in Germany: A Multicenter study. Clin Infect Dis. 2013 2013-04-15;56(8):1132–1140.
- Croxen MA, Law RJ, Scholz R, et al. Recent Advances in Understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013 2013-10-01;26(4):822–880.
- Venegas-Vargas C, Henderson S, Khare A, et al. Factors associated with Shiga toxin-producing Escherichia coli shedding by Dairy and beef cattle [Journal article; research Support, Non-U.S. Gov't; research Support, N.I.H., Extramural; research Support, U.S. Gov't, Non-P.H.S.]. Appl Environ Microbiol. 2016 2016-08-15;82(16):5049–5056.
- Baschera M, Cernela N, Stevens MJA, et al. Shiga toxin-producing Escherichia coli (STEC) isolated from fecal samples of African dromedary camels. One health. 2019 Jun;7, 100087:1-5.
- Paton JC, Paton AW. Pathogenesis and Diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998 1998-01-01;11(3):450–479.
- Melton-Celsa AR. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiology Spectrum. 2014, 2014-08-22;2(4):1-13.
- Girard F, Batisson I, Frankel GM, et al. Interaction of enteropathogenic and Shiga toxin-producing Escherichia coli and porcine intestinal mucosa: role of intimin and Tir in adherence [Journal article; research Support, Non-U.S. Gov't]. Infect Immun. 2005 2005-09-01;73(9):6005–6016.
- Lai Y, Rosenshine I, Leong JM, et al. Intimate host attachment: enteropathogenic and enterohaemorrhagic Escherichia coli. Cellular Microbiology. 2013, 5 (11):1796-1808.
- Oswald E, Schmidt H, Morabito S, et al. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant [Journal article; research Support, Non-U.S. Gov't]. Infect Immun. 2000 2000-01-01;68(1):64–71.
- Battle SE, Brady MJ, Vanaja SK, et al. Actin Pedestal formation by Enterohemorrhagic Escherichia coli Enhances bacterial host cell Attachment and Concomitant type III Translocation. Infect Immun. 2014;82(9):3713–3722.
- Shaw RK, Cleary J, Murphy MS, et al. Interaction of enteropathogenic Escherichia coli with human intestinal mucosa: role of effector proteins in brush border remodeling and formation of attaching and effacing lesions [Journal article; research Support, Non-U.S. Gov't]. Infect Immun. 2005 2005-02-01;73(2):1243–1251.
- Luo Y, Frey EA, Pfuetzner RA, et al. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex [Journal article; research Support, Non-U.S. Gov't]. Nature. 2000 2000-06-29;405(6790):1073–1077.
- Furniss RCD, Clements A. Regulation of the Locus of Enterocyte Effacement in Attaching and Effacing Pathogens. Journal of Bacteriology. 2018 2017-07-31;200(2):e00336-17.
- Kelly G, Prasannan S, Daniell S, et al. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli [Journal article; research Support, Non-U.S. Gov't]. Nat Struct Biol. 1999 1999-04-01;6(4):313–318.
- Yang X, Sun H, Fan R, et al. Genetic diversity of the intimin gene (eae) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Scientific Reports. 2020;10(1):3275-3283.
- Zhang WL, Kohler B, Oswald E, et al. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains [Journal article; research Support, Non-U.S. Gov't]. J Clin Microbiol. 2002 2002-12-01;40(12):4486–4492.
- Fierz L, Cernela N, Hauser E, et al. Characteristics of Shiga toxin-Producing Escherichia coli Strains Isolated during 2010–2014 from Human Infections in Switzerland. Frontiers in Microbiology. 2017, 3(8):1471-1477.
- Blanco M, Schumacher S, Tasara T, et al. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-eta2). BMC Microbiol. 2005 May 9;5:23.
- Xu Y, Bai X, Zhao A, et al. Genetic diversity of intimin gene of atypical Enteropathogenic Escherichia coli isolated from human, animals and Raw Meats in China. PLOS ONE. 2016 2016-03-31;11(3):e0152571.
- Matussek A, Jernberg C, Einemo IM, et al. Genetic makeup of Shiga toxin-producing Escherichia coli in relation to clinical symptoms and duration of shedding: a microarray analysis of isolates from Swedish children. Eur J Clin Microbiol Infect Dis. 2017;36(8):1433–1441.
- Lagerqvist N, Lof E, Enkirch T, et al. Outbreak of gastroenteritis highlighting the diagnostic and epidemiological challenges of enteroinvasive Escherichia coli, County of Halland, Sweden, November 2017 [Journal Article]. Euro Surveill. 2020-03-01;25(9): 1900466.
- Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biology. 2018;19(1):153-165.
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: A New genome Assembly Algorithm and Its Applications to Single-cell sequencing. J Comput Biol. 2012;19(5):455–477.
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014 2014-07-15;30(14):2068–2069.
- Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications [Journal article; research Support, N.I.H., Extramural]. BMC Bioinform. 2009 2009-12-15;10:421.
- De Rauw K, Buyl R, Jacquinet S, et al. Risk determinants for the development of typical haemolytic uremic syndrome in Belgium and proposition of a new virulence typing algorithm for Shiga toxin-producing Escherichia coli. Epidemiology and Infection. 2019;147:1-5.
- Karmali MA. Factors in the emergence of serious human infections associated with highly pathogenic strains of shiga toxin-producing Escherichia coli. Int J Med Microbiol. 2018;308(8):1067–1072.
- Mora A, López C, Dhabi G, et al. Seropathotypes, Phylogroups, Stx subtypes, and intimin types of Wildlife-carried, Shiga toxin-producing Escherichia coli strains with the same characteristics as human-pathogenic isolates. Appl Environ Microbiol. 2012 2012-04-15;78(8):2578–2585.
- Preussel K, Hohle M, Stark K, et al. Shiga toxin-producing Escherichia coli O157 is more likely to lead to hospitalization and death than non-O157 serogroups–except O104 [Comparative study; Journal article]. PLoS One. 2013 2013-01-20;8(11):e78180.
- Beutin L, Krause G, Zimmermann S, et al. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period [Journal article; Multicenter study; research Support. Non-U.S. Gov't]. J Clin Microbiol. 2004 2004-03-01;42(3):1099–1108.
- Ramachandran V, Brett K, Hornitzky MA, et al. Distribution of intimin subtypes among Escherichia coli isolates from ruminant and human sources. J Clin Microbiol. 2003 Nov;41(11):5022–5032.
- Tostes R, Goji N, Amoako K, et al. Subtyping Escherichia coli virulence genes isolated from Feces of beef cattle and clinical Cases in Alberta. Foodborne Pathog Dis. 2017;14(1):35–42.
- Kozub-Witkowski E, Krause G, Frankel G, et al. Serotypes and virutypes of enteropathogenic and enterohaemorrhagic Escherichia coli strains from stool samples of children with diarrhoea in Germany. J Appl Microbiol. 2008 Feb;104(2):403–410.
- Feng PC, Keys C, Lacher D, et al. Prevalence, characterization and clonal analysis of Escherichia coli O157: non-H7 serotypes that carry eae alleles. FEMS Microbiol Lett. 2010 Jul 1;308(1):62–67.
- Brooks JT, Sowers EG, Wells JG, et al. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002 [Journal article]. J Infect Dis. 2005 2005-10-15;192(8):1422–1429.
- Werber D, Fruth A, Buchholz U, et al. Strong association between Shiga toxin-producing Escherichia coli O157 and virulence genes stx 2 and eae as Possible Explanation for Predominance of Serogroup O157 in patients with Haemolytic Uraemic syndrome. Eur J Clin Microbiol Infect Dis. 2003 2003-12-01;22(12):726–730.
- Ethelberg S, Olsen KEP, Scheutz F, et al. Virulence Factors for hemolytic uremic syndrome, Denmark1. Emerging Infect. Dis.. 2004 2004-01-01;10(5):842–847.
- Mody RK, Griffin PM. Editorial Commentary: fecal shedding of Shiga toxin-producing Escherichia coli: What should Be Done to Prevent secondary Cases? Clin Infect Dis. 2013 2013-04-15;56(8):1141–1144.