ABSTRACT
We report surveillance results of Cache Valley virus (CVV; Peribunyaviridae, Orthobunyavirus) from 2017 to 2020 in New York State (NYS). Infection rates were calculated using the maximum likelihood estimation (MLE) method by year, region, and mosquito species. The highest infection rates were identified among Anopheles spp. mosquitoes and we detected the virus in Aedes albopictus for the first time in NYS. Based on our previous Anopheles quadrimaculatus vector competence results for nine CVV strains, we selected among them three stains for further characterization. These include two CVV reassortants (PA and 15041084) and one CVV lineage 2 strain (Hu-2011). We analyzed full genomes, compared in vitro growth kinetics and assessed vector competence of Aedes albopictus. Sequence analysis of the two reassortant strains (PA and 15041084) revealed 0.3%, 0.4%, and 0.3% divergence; and 1, 10, and 6 amino acid differences for the S, M, and L segments, respectively. We additionally found that the PA strain was attenuated in vertebrate (Vero) and mosquito (C6/36) cell culture. Furthemore, Ae. albopictus mosquitoes are competent vectors for CVV Hu-2011 (16.7–62.1% transmission rates) and CVV 15041084 (27.3–48.0% transmission rates), but not for the human reassortant (PA) isolate, which did not disseminate from the mosquito midgut. Together, our results demonstrate significant phenotypic variability among strains and highlight the capacity for Ae. albopictus to act as a vector of CVV.
Introduction
Cache Valley virus (CVV; Peribunyaviridae, Orthobunyavirus) is an emerging mosquito-borne pathogen endemic to North America [Citation1]. The CVV genome is a negative sense, single stranded RNA organized into three distinct segments designated L (large), M (medium) and S (small). The L segment encodes the RNA-dependent RNA polymerase; the M segment encodes two glycoproteins (Gn and Gc) and a non-structural protein (NSm); and the S segment encodes the nucleocapsid protein (N) and a second non-structural protein (NSs) [Citation2,Citation3].
CVV is endemic to Canada, Mexico, and the United States, where the virus circulates in mosquitoes and mammals including sheep, cattle and white-tailed deer [Citation4]. With prevalence as high as 69% in livestock reported in some regions, CVV is an important cause of embryonic and fetal death or neonatal malformations, resulting in significant economic losses [Citation5–7]. Despite its importance in the livestock industry, no vaccinations or treatments are available. Humans are considered dead-end hosts and while reported human disease is rare, CVV has been associated with neuroinvasive illness [Citation8–11]. Although recent widespread serosurveys are lacking, historic reports suggest seroprevalence may be as high as 40% in some locations in the U.S. [Citation1].
CVV isolates are grouped into 2 lineages [Citation4]. Since 2010 Lineage 2 was shown to have displaced lineage 1 in Connecticut, New York, and Canada [Citation4,Citation12]. Our previous studies demonstrate that Anopheles (An.) quadrimaculatus have increased competence for lineage 2 CVV strains, which likely contributed to the displacement and increased prevalence of CVV in the region over the last decade. In addition, we previously found evidence of segment reassortment in recent strains [Citation12]. For bunyaviruses, reassortments of genome segments during co-infection has played an important role in generating diversity that can confer important alterations to viral fitness and transmissibility in hosts and vectors [Citation13–17].
In order to expand on previous results, we report surveillance testing of CVV from 2017 to 2020 in New York State (NYS). In addition, we determined vector competence of Aedes (Ae.) albopictus for current genetically and phenotypically distinct circulating strains of CVV, including: one lineage 2 strain isolated from a human and two reassortant strains isolated from a human and a pool of mosquitoes. Our results further demonstrate the influence of CVV genetics on transmission and highlight the increasing threat of Ae. albopictus as a vector of endemic viruses.
Materials and methods
Mosquito surveillance
Adult mosquitoes were collected from 2017 to 2020 in the following regions in NYS: West, Finger Lakes, North, Central, Hudson Valley and Long Island, using CDC light traps baited with CO2 [Citation18] or gravid traps [Citation19]. Mosquitoes were identified to species morphologically [Citation20], and females were pooled in groups of 4–50 individuals by trap type, date collected and trap location. Pools were transported on dry ice to the Arbovirus Laboratories, Wadsworth Center, New York State Dept Health, for testing and were stored at −80°C until processed.
Mosquito pools were processed as previously described [Citation21]. Briefly, they were homogenized in 1 ml of mosquito diluent (MD) containing 20% fetal bovine serum, 50 μg of streptomycin per mL, 50 U of penicillin, and 2.5 μg of amphotericin B per mL in phosphate-buffered saline, in a Retsch Mixer Mill set to 24 cycles/s for 2 min. The tubes were then centrifuged for 8 min at 12,000 rpm and the supernatant removed. Approximately 500 μl of supernatant was frozen at −80°C for storage while the remainder was used for RNA extraction [Citation22].
Extraction plates (Thermofisher, Waltham, MA, USA) were prepared on a Tecan Evo 150 liquid handler (Tecan, Morrisvelle, NC, USA) and 50 μL of homogenates were used to extract RNA on a Magmax 96 Express (Applied Biosystems, Waltham, MA, USA) using a MagMax viral isolation kit (Thermofisher, Waltham, MA, USA). A total of 90 μL of homogenized sample RNA was eluted. Real- time RT–PCR assay was performed by using CVV_F (ACAGCCAATGGTGTCGAAAAC), CVV_R (TGCAGGGATGCTAGACAAGATG) primers, and CVV_P (6FAM-CTGACGGTATTGAATCAGCAT-MGBNFQ) probe for CVV detection. Maximum likelihood estimation (MLE) calculations to determine prevalence of CVV in mosquitoes were based upon a program developed by Dr. Brad Biggerstaff as shown on the CDC website, https://www.cdc.gov/westnile/resourcepages/mosqsurvsoft.html [Citation23,Citation24]. Statistical analysis of the virus detection data by year, species, and regions (number of pools) were carried out at a significance level of p < 0.05 using OpenEpi, Version 3, open-source calculator–TwobyTwo (https://www.openepi.com/TwobyTwo/TwobyTwo.htm). In addition, we compared the relative mosquitoes abundance between years, regions and the positive mosquito species using a Poisson regression model.
Genetic analyses of Cache Valley virus
The full genome sequences of two CVV human strains (the lineage 2 Hu-2011 strain isolated from cerebrospinal fluid and the reassortant PA strain isolated from brain tissue) [Citation9,Citation10] and a reassortant isolated from An. quadrimaculatus (15041084) obtained from our earlier study were analysed [Citation12]. The two CVV reassortant strains (PA and 15041084) contain lineage 1 L segment and lineage 2 S and M segments and both come from counties in western New York [Citation12]. In addition, PA and 15041084 strains presented different phenotypes in An. quadrimaculatus infectivity [Citation12]. Nucleotide and amino acid sequence alignments were created with CVV coding regions by using the MegAlign module of the DNAStar software package (https://www.dnastar.com).
Viral growth kinetics
Confluent monolayers of each cell type in six-well plates were infected in duplicate at a multiplicity of infection (MOI) of 0.01 plaque forming units (pfu) per cell with 100 μl of each virus, and incubated for 1 h at 37°C or 28°C for African Green Monkey kidney (Vero) and Ae. albopictus cells (C6/36), respectively, with 5% CO2. The inoculum was removed, and cells were washed twice with BA-1 (blood agar media) to remove any remaining virus. Infected plates were then overlaid with 3 ml maintenance media (Eagle minimum essential medium with 2% fetal bovine serum heat inactivated with ½ g/L sodium bicarbonate plus 0.1 mM non-essential amino acids plus 100 U/ml penicillin–streptomycin). Supernatant (100 μl) was sampled at 24 h intervals up to 96 h post-infection (4 days), 20% FBS were added to each sample and stored at −80°C. Titrations were performed in duplicate by plaque assay on Vero cells [Citation25] and mean titres for each time point were calculated. Growth kinetics were compared using repeated measured ANOVA and Tukey’s post hoc tests (GraphPad Prism, Version 5.0) [Citation26].
Mosquito vector competence
Ae. albopictus (Spring Valley, NY, USA; kindly provided by Laura Harrington, Cornell University) were established in 2019 from field-collected eggs. Eggs were hatched in distilled water and maintained under standard rearing conditions (27 ± 1°C; 70% relative humidity; 12:12-h light:dark photoperiod) [Citation25,Citation27]. F7 females were used for the CVV experiments.
Two CVV strains isolated in humans (Hu-2011and PA) and the reassortant 15041084, isolated in mosquitoes [Citation9,Citation10] were used to infect Vero cells at a MOI of 0.01 pfu/cell using 100 μl of each virus and maintained at 37°C, 5% CO2. At 48 h following infection, the supernatants were harvested and diluted 1:1 with defibrinated sheep blood plus a final concentration of 2.5% sucrose. In addition, fresh virus supernatants were diluted to 1:100 (Hu-2011) or 1:1000 (15041084) in C6/36 maintenance media before being mixed 1:1 with defibrinated sheep blood with a final concentration of 2.5% sucrose. Only the PA strain supernatants were not diluted before being mixed with defibrinated sheep blood. Female Ae. albopictus (5–7 days old) deprived of sugar for 24 h were allowed to feed on the infectious bloods for 45 min via a Hemotek membrane feeding system (Discovery Workshops, Acrington, UK) with a porcine sausage casing membrane at 37°C [Citation28]. Following feeding, females were anesthetized with CO2 and fully engorged mosquitoes were transferred to 0.6 L cardboard containers and maintained with 10% sucrose under standard rearing conditions [Citation27] until harvested for testing. A 1 ml aliquot of each blood meal pre-feeding was frozen at −80°C and CVV titres were determined by plaque assay on Vero cells [Citation25]. Three biological replicates were performed for each CVV strain.
Infection, dissemination, and transmission assays were performed on days 7 and 14 post infectious blood meal as previously described [Citation28]. Dissemination rate is the proportion of mosquitoes with infected legs among the infected mosquitoes. Transmission rate is the proportion of mosquitoes with positive saliva among mosquitoes with disseminated infection. Real-time RT–PCR was used to detect CVV following sample processing as previously described [Citation28]. A Fisher’s exact test was used to compare mosquito infection rates, dissemination rates, and transmission rates between groups exposed to distinct CVV isolates at both time points. All statistical analyses were carried out at a significance level of p < 0.05 using OpenEpi, Version 3, open source calculator–TwobyTwo (https://www.openepi.com/TwobyTwo/TwobyTwo.htm).
Results
We tested 441,139 female mosquitoes in 13,258 mosquito pools from 2017 to 2020, yielding a total of 72 CVV positive pools. Comparisons of MLE of prevalence were made by year (A), region (B), and mosquito species (C). CVV activity was detected during each of the 4 years studied with the highest estimates of prevalence in 2017 (0.38, 95% CI [0.29, 0.51]) and 2020 (0.17, 95% CI [0.09, 0.29]) (A). Except for 2019 vs 2020, significant differences in prevalence were observed between years studied (Fisher’s exact test, P < 0.01, OR: 5.55, 95% CI: 1.211–51.6) (A). With the exception of the North region, CVV was detected throughout NYS, with the highest prevalence in the Finger Lakes (0.46, 95% CI [0.08, 1.49]) (B). However, Long Island was the only region where the prevalence of CVV pools showed a significant difference compared to Finger Lakes (Fisher’s exact test, P < 0.01, OR: 16.48, 95% CI: 1.186–227.9). No significant differences in CVV prevalence was identified for the Central and West regions, nor the Hudson Valley and Long Island regions (B). The virus was detected from nine mosquito species, with the highest prevalence in An. punctipennis (1.12, 95% CI [0.55, 2.05]), followed by Ae. cinereus (0.75, 95% CI [0.2, 2.03]), An. quadrimaculatus (0.64, 95% CI [0.3, 1.2]) and Coquillettidia perturbans, (0.29, 95% CI [0.21, 0.39]) with no significant differences in CVV prevalence measured between these four species (C). Furthermore, the only significant interaction effects were measured when we considered interactions of the relative mosquito abundance between year and region with increase of mosquito abundance in the Finger Lakes in 2019 (p = 0.0025), in Long Island in 2019 (p = 1.93e−07) and in the West in 2019 (p = 0.0155), as compare to Central region in 2017.
Figure 1. Cache Valley virus prevalence in New York State. Infection rates were calculated by Maximum Likelihood Estimation (MLE) for (A) year (Fisher’s exact test, ***P < 0.01, OR: 5.55, 95% CI: 1.211–51.6), (B) New York State region (Fisher’s exact test, ***P < 0.01, OR: 16.48, 95% CI: 1.186–227.9), (C) mosquito species (Fisher’s exact test, ***P < 0.003, OR: 0.05, 95% CI: 0.004–0.453), or (D) year for Ae. albopictus. Bars represent upper and lower limits of infection rate based on 95% confidence levels. Values on top represent number of pools tested. MLEs were calculated using https://www.cdc.gov/westnile/resourcepages/mosqsurvsoft.html. ***P < 0.05.
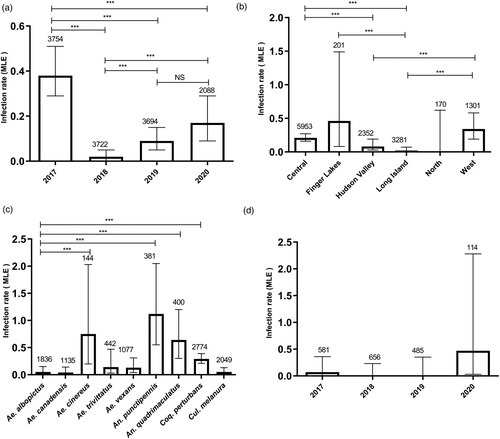
Ae. albopictus accounted for 9.8% of the total mosquitoes collected (N = 43,267, 1836 pools) during the study period (D). The invasive mosquito was collected in three different regions including Hudson Valley (11,477 mosquitoes), Long Island (31,779 mosquitoes) and, for the first time, in Northern NYS (11 mosquitoes) in 2020. CVV was detected in 2 pools of Ae. albopictus (0.05, 95% CI [0.01, 0.15]) in NYS (Long Island in 2017 and Hudson Valley in 2020) with a similar prevalence as Ae. canadensis (0.04, 95% CI [0.01, 0.14]), Ae. trivittatus (0.14, 95% CI [0.03, 0.47]), Ae. vexans (0.13, 95% CI [0.04, 0.31]) and Culiseta melanura (0.05, 95% CI [0.01, 0.13]) (C). This represents the first isolation of CVV from Ae. albopictus in NYS.
Sequence analysis of the two reassortant strains (PA and 15041084) revealed 0.3%, 0.4%, 0.3% nucleotide divergence and 1, 10, and 6 amino acid differences for the S, M, and L segments, respectively. In addition, the PA and Hu-2011 (Lineage 2) strains showed 0.5%, 0.7% and 6.3% nucleotide divergence, and 1, 12, and 29 amino acid difference for the S, M, and L segments, respectively (). For the S segment, the only unique amino acid, F70S, occurred in the PA strain NSs protein. For the M segment, the NSm protein of 15041084 and Hu-2011 strains are identical and differed from the PA strain by two amino acids, V371G and K450Q. The Gc protein of the PA strain was found to be more divergent, with 8 and 10 amino acid differences relative to the 15041084 and Hu-2011strains, respectively. With the exception of amino acid changes T666A and V845D, the Gc protein of the 15041084 and Hu-2011 strains were identical (). Consistent with other lineage 2 strains, the L protein of the Hu-2011strain was most distinct from the reassortant strains, with 27 and 29 amino acid differences in comparison with the 15041084 and PA strains, respectively ().
Table 1. Amino acid differences among Cache Valley virus (CVV) strains used for vector competence and growth kinetic studies.
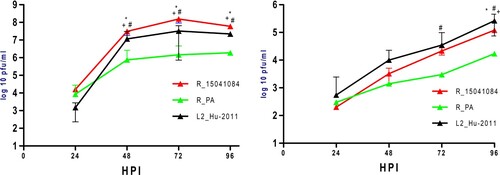
Growth kinetics of PA, 15041084 and Hu-2011 were compared on Vero (37°C) and C6/36 cells (28°C) (). The 15041084 strain replicated to significantly higher titres than the two other CVV strains at 48, 72 and 96 h on Vero cells (t-test, p < 0.05) with ∼8 log10 PFU/mL peak viral titre observed at 72 h (A). With the exception of 24 h, significant differences were also identified between Hu-2011 (∼7 log10 PFU/mL peak viral titre at 72 h) and PA (∼6 log10 PFU/mL peak viral titre at 96 h) strains at all other time points on Vero cells (t-test, p < 0.05; A). In C6/36 cells, no significant differences were observed at 24 and 48 h between strains. However, the PA strain showed significantly lower titres as compared to 15041084 at 96 h, and Hu-2011 at 72 and 96 h (t-test, p < 0.05; B).
Figure 2. Growth kinetics of unique Cache Valley virus strains in cell culture. Kinetics were determined in (A) mammalian (Vero) or (B) mosquito (C6/36) cells. Points represent means of duplicate values ± standard deviation. #Denotes significant difference (Two-way ANOVA, p < 0.001 by Tukey post hoc test) between PA and Hu-2011 CVV strains at indicated time points in both cell lines. *Denotes significant difference (Two-way ANOVA, p < 0.001 by Tukey post hoc test) between Hu-2011 and 15041084 CVV strains at indicated time points in both cell lines. +Denotes significant difference (Two-way ANOVA, p < 0.001 by Tukey post hoc test) between PA and 15041084 CVV strains at indicated time points in both cell lines. A significant effect independent of the time was measured in both cell lines (Two-way ANOVA, p < 0.001).
Vector competence assays with Ae. albopictus for CVV Hu-2011, PA, and 15041084 were conducted to determine the transmission potential of Ae. albopictus for genetically distinct CVV strains (). When mosquitoes fed on high virus titres (≥5.5 log10PFU/mL), infection rates were 100.0% for all three CVV isolates at 14 days post-infection (dpi). Dissemination rates for both Hu-2011 and 15041084 were also 100% yet, strikingly, mosquitoes exposed to CVV PA did not show evidence of dissemination. Transmission rates for CVV Hu-2011 and CVV 15041084 were also high, 61.9% and 48.0%, respectively (). Significant reduction in infection rates were observed following exposure to blood meals with lower input titres for CVV 15041084 (4.2 to 3.6 log10 PFU/mL) (Fisher’s exact test, P < 0.001, OR: 32.5, 95% CI: 6.675–175.4; P < 0.001, OR: 6.538, 95% CI: 1.739-27.14) and the lineage 2 CVV Hu-2011 (5 to 4.5 log10 PFU/mL; Fisher’s exact test, P < 0.001, OR: 37.92, 95% CI: 4.671–1638; P < 0.001, OR: 43.5, 95% CI: 5.322–1872) at 7 and 14 dpi, respectively. However, no significant differences in the dissemination rates were identified for CVV 15041084 at 7 and 14 dpi. In addition, exposure to lower blood meal titres significantly reduced CVV Hu-2011 dissemination rate at 7 dpi (Fisher’s exact test, P < 0.02, OR: 12.89, 95% CI: 1.023–655.8) and transmission rates at 14 dpi (Fisher’s exact test, P < 0.009, OR: 8.182, 95% CI: 1.304–85.84). Furthermore, between 7 and 14 dpi significant increases in transmission rates were observed for the CVV 15041084 (P < 0.04, OR: 0.2424, 95% CI: 0.03745–1.209) and CVV Hu-2011 (P < 0.001, OR: 0.1528, 95% CI: 0.03948-0.5573). Despite this, transmission was measured for both strains at the lowest doses utilized (). Our results indicate that Ae. albopictus is a competent vector for CVV Hu-2011 and the CVV 15041084, but not for the human reassortant (PA) isolate, which did not disseminate from the mosquito midgut ().
Table 2. Infection, dissemination and transmission rates of Ae. albopictus following exposure to distinct Cache Valley virus (CVV) strains.
Discussion
Our study confirmed yearly variability in CVV activity in NYS and association of the virus with various mosquito genera including Aedes, Anopheles, Coquillettidia and Culiseta, as described previously [Citation12,Citation29]. In addition, our results corroborate previous findings showing higher prevalence of CVV in An. punctipennis and An. quadrimaculatus [Citation12,Citation29]. Notably, CVV was detected for the first time in Ae. albopictus collected in NYS, suggesting the potential involvement of this species in the transmission cycle. Previous isolations of CVV from Ae. albopictus have been reported in Connecticut and New Jersey [Citation4,Citation30]. Interestingly, all CVV isolates from Ae. albopictus in northeastern USA belong to lineage 2 [Citation31].
The Asian tiger mosquito (Ae. albopictus) is a highly invasive species that has been introduced into the U.S. and has become permanently established in at least 27 states, including NYS [Citation32]. Our retrospective surveillance data in NYS from 2000 to 2016 confirmed that Ae. albopictus populations are well established in the Long Island and Hudson Valley regions [Citation12,Citation33]; however, the first detection in the northern NYS suggests that this species may be continuing to expand its range in the northeastern U.S. The distribution and spread of this species is coincidental with the high prevalence of CVV in white-tailed deer in NYS [Citation34]. However, many factors, including mosquito, host feeding preference (mammalian) and viral evolution, could be contributing to the increase of CVV circulation in the region.
Previous studies have shown that both the M and L segments of bunyaviruses have a role in determining host virulence and neuropathogenicity, as well as mosquito infectivity [Citation35–39]. We previously demonstrated increased infectivity of lineage 2 CVV strains in Anopheles, consistent with increased prevalence following lineage 1 displacement in the Northeast [Citation12,Citation29]. In addition, the three CVV reassortant mosquito isolates that contained only the lineage 1 L segment were more infectious than other lineage 1 strains, suggesting a role for the S and/or M segments in the increased mosquito infectivity of lineage 2 strains [Citation12]. However, the human reassortant strain (PA) sharing the lineage 1 L segment and the lineage 2 S and M segments with the mosquito reassortant strains was not transmitted by An. quadrimaculatus [Citation12]. The high susceptibility of Ae. albopictus to CVV obtained in our results are similar to the reports of others [Citation31,Citation40]. In this study, using Ae. albopictus mosquitoes, we confirmed the PA phenotype observed previously with An. quadrimaculatus. We additionally demonstrated that this strain was attenuated in vertebrate and invertebrate cell culture. Together, our data suggest that individual mutations, in addition to segment reassortment, can play a critical role in determining CVV fitness.
Our sequence analysis revealed amino acid difference in NSs (1aa), NSm (2 aa), Gc (8 aa) and L (6 aa) proteins between the two reassortant strains (PA and 15041084). Previous studies showed that NSs and NSm proteins are not essential for bunyaviruses viability but deletions of the NSs proteins reduced viral replication [Citation41–43]. Furthermore, the NSs protein is a major virulence factor and plays an important role in viral evasion of innate immunity [Citation41,Citation42]. Rift Valley Fever virus (RVFV) NSs has been shown to influence dissemination rates in Ae. aegypti while deletion of the RVFV NSm was sufficient to nearly abolish mosquito infection [Citation44,Citation45]. Moreover, for orthobunyaviruses it is generally accepted that both glycoproteins are required for virus entry. Although little is known about individual Gn and Gc protein functions, it is suggested that Gc is the attachment protein for mammalian and mosquito cells [Citation46–50]. For La Crosse virus and California encephalitis virus, the specificity of virus-vector interactions is thought to be strongly influenced by the efficiency of the fusion function of the Gc (G1) envelope glycoprotein operating at the midgut level in the arthropod vector [Citation46,Citation50]. Understanding of the biological function of the non-structural and Gc proteins of CVV is critical to uncovering the role of individual mutations in host-specific fitness and transmissibility of emergent CVV strains.
The invasiveness of Ae. albopictus, as well his zoophilic behaviour with a preference for human blood [Citation32] and its potential to transmit endemic and invasive arboviruses, could increase the threat from local and introduced viruses in the Northeast U.S. Further, the general increase in CVV activity, the capacity for CVV transmission, and the influence of viral genetics on vector competence, suggest Ae. albopictus are likely to contribute to the expanding threat of CVV transmission and disease.
Acknowledgements
We would like to thank the New York State Bureau of Communicable Disease Control and the NYS county health departments for mosquito collections and data entry. We thank the New York State Department of Health, Wadsworth Center Media and Tissue Core Facility for providing cells and media for these studies and the Wadsworth Center Applied Genomics Technologies Core for viral sequencing. We additionally thank the NYS Arbovirus Laboratory insectary staff for assistance with rearing and experimentation. Technical assistance was also provided by Maya Andonova and Kimberly Holloway.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Waddell L, Pachal N, Mascarenhas M, et al. Cache Valley virus: a scoping review of the global evidence. Zoonoses Public Heal. 2019;66:739–758.
- Dunlop JI, Szemiel AM, Navarro A, et al. Development of reverse genetics systems and investigation of host response antagonism and reassortment potential for Cache Valley and Kairi viruses, two emerging orthobunyaviruses of the Americas. PLoS Negl Trop Dis. 2018;12:e0006884.
- Elliott RM. Molecular biology of the Bunyaviridae. J Gen Virol. 1990;71:501–522.
- Armstrong PM, Andreadis TG, Anderson JF. Emergence of a new lineage of Cache Valley virus (Bunyaviridae : Orthobunyavirus) in the Northeastern United States. Am J Trop Med Hyg. 2015;93:11–17.
- Chung SI, Livingston CWJ, Jones CW, et al. Cache Valley virus infection in Texas sheep flocks. J Am Vet Med Assoc United States. 1991;199:337–340.
- Meyers MT, Bahnson CS, Hanlon M, et al. Management factors associated with operation-level prevalence of antibodies to Cache Valley virus and other Bunyamwera serogroup viruses in sheep in the United States. Vector-Borne Zoonotic Dis. 2015;15:683–693.
- Uehlinger FD, Wilkins W, Godson DL, et al. Seroprevalence of Cache Valley virus and related viruses in sheep and other livestock from Saskatchewan, Canada. Can Vet J. 2018;59:413–418.
- Campbell GL, Mataczynski JD, Reisdorf ES, et al. Second human case of Cache Valley virus disease. Emerg Infect Dis. 2006;12:854–856.
- Nguyen NL, Zhao G, Hull R, et al. Cache Valley virus in a patient diagnosed with aseptic meningitis. J Clin Microbiol. 2013;51:1966–1969.
- Yang Y, Qiu J, Snyder-Keller A, et al. Fatal Cache Valley virus meningoencephalitis associated with rituximab maintenance therapy. Am J Hematol. 2018;93:590–594.
- Wilson MR, Suan D, Duggins A, et al. A novel cause of chronic viral meningoencephalitis: Cache Valley virus. Ann Neurrology. 2017;82:105–114.
- Dieme C, Ngo KA, Tyler S, et al. Role of Anopheles mosquitoes in Cache Valley virus lineage displacement, New York, USA. Emerg Infect Dis. 2022;28:303–313.
- Horne KME, Vanlandingham DL. Bunyavirus-vector interactions. Viruses. 2014;6:4373–4397.
- Elliott RM. Bunyaviruses and climate change. Clin Microbiol Infect European Soc Clinic Microbiol Infect Dis. 2009;15:510–517.
- Pringle CR. Genetics and genome segment reassortment. In: Elliott RM, editor. The Bunyaviridae. The Viruses. Boston (MA): Springer; 1996. DOI:10.1007/978-1-4899-1364-7_8
- Mcdonald SM, Nelson MI, Turner PE, et al. Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat Rev Microbiol. 2016;14:448–460.
- Vijaykrishna D, Mukerji R, Smith GJD. RNA virus reassortment: an evolutionary mechanism for host jumps and immune evasion. PLoS Pathog. 2015;11:1–6.
- Newhouse VF, Siverly RE. St. Louis encephalitis virus from mosquitoes in southwestern Indiana, 1964. J Med Entomol. England. 1966;3:340–342.
- Reiter P. A portable battery-powered trap for collecting gravid Culex mosquitoes. Mosq News. 1983;43:496–498.
- Means RG. Mosquitoes of New York, part II: genera of culicidae other than Aedes occuring in New York. New York State Museum. 1987;430B:1–180.
- Oliver J, Lukacik G, Kokas J, et al. Twenty years of surveillance for Eastern equine encephalitis virus in mosquitoes in New York State from 1993 to 2012. Parasit Vecs. 2018;11:362.
- Kauffman EB, Jones SA, Dupuis AP, et al. Virus detection protocols for West Nile virus in vertebrate and mosquito specimens. J Clin Microbiol. 2003;41:3661–3667.
- Bialosuknia SM, Tan Y, Zink SD, et al. Evolutionary dynamics and molecular epidemiology of west Nile virus in New York state: 1999–2015. Virus Evol. 2019;5:1–13.
- Eastwood G, Shepard JJ, Misencik MJ, et al. Local persistence of novel regional variants of La Crosse virus in the Northeast USA. Parasit Vec BioMed Central. 2020;13:1–8.
- Dieme C, Ciota AT, Kramer LD. Transmission potential of Mayaro virus by Aedes albopictus, and Anopheles quadrimaculatus from the USA. Parasit Vec BioMed Central. 2020;13:4–9.
- Kuo L, Jaeger AS, Banker EM, et al. Reversion to ancestral Zika virus NS1 residues increases competence of Aedes albopictus. PLoS Pathog. 2020;16:1–16.
- Ciota AT, Matacchiero AC, Kilpatrick AM, et al. The effect of temperature on life history traits of Culex mosquitoes. J Med Entomol. 2014;51:55–62.
- Ciota AT, Bialosuknia SM, Zink SD, et al. Effects of Zika virus strain and Aedes mosquito species on vector competence. Emerg Infect Dis. 2017;23:1110–1117.
- Andreadis TG, Armstrong PM, Anderson JF, et al. Spatial-temporal analysis of Cache Valley virus (Bunyaviridae: Orthobunyavirus) infection in anopheline and culicine mosquitoes (Diptera: Culicidae) in the Northeastern United States, 1997–2012. Vector-Borne Zoonotic Dis. 2014;14:763–773.
- Armstrong PM, Anderson JF, Farajollahi A, et al. Isolations of Cache Valley virus from Aedes albopictus (Diptera : Culicidae) in New Jersey and evaluation of its role as a regional arbovirus vector. J Med Entomol. 2013;50:1310–1314.
- Ayers VB, Huang YJS, Lyons AC, et al. Infection and transmission of Cache Valley virus by Aedes albopictus and Aedes aegypti mosquitoes. Parasit Vectors. 2019;12:384.
- Kraemer MUG, Reiner RC, Brady OJ, et al. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat Microbiol. 2019;4:854–863.
- Shragai T, Harrington LC. Aedes albopictus (Diptera: Culicidae) on an invasive edge: abundance, spatial distribution, and habitat usage of larvae and pupae across urban and socioeconomic environmental gradients. J Med Entomol. 2019;56:472–482.
- Dupuis AP, Prusinski MA, Russell A, et al. Serologic survey of mosquito-borne viruses in hunter-harvested white-tailed deer (odocoileus virginianus), New York State. Am J Trop Med Hyg. 2020;104:593–603.
- Hughes HR, Lanciotti RS, Blair CD, et al. Full genomic characterization of California serogroup viruses, genus Orthobunyavirus, family Peribunyaviridae including phylogenetic relationships. Virology. 2017;512:201–210.
- Endres MJ, Griot C, Gonzalez-Scarano F, et al. Neuroattenuation of an avirulent bunyavirus variant maps to the L RNA segment. J Virol. 1991;65:5465–5470.
- Briese T, Calisher CH, Higgs S. Viruses of the family Bunyaviridae: are all available isolates reassortants? Virology. 2013;446:207–216.
- Cheng LL, Rodas JD, Schultz KT, et al. Potential for evolution of California serogroup bunyaviruses by genome reassortment in Aedes albopictus. Am J Trop Med Hyg. 1999;60:430–438.
- Fausta Dutuze M, Nzayirambaho M, Mores CN, et al. A review of Bunyamwera, Batai, and Ngari viruses: understudied Orthobunyaviruses with potential one health implications. Front Vet Sci. 2018;5:1–9.
- Chan KK, Auguste AJ, Brewster CC, et al. Vector competence of Virginia mosquitoes for Zika and Cache Valley viruses. Parasit Vec BioMed Central. 2020;13:1–9.
- Eifan S, Schnettler E, Dietrich I, et al. Non-structural proteins of arthropod-borne bunyaviruses: roles and functions. Viruses. 2013;5:2447–2468.
- Elliott RM. Orthobunyaviruses: recent genetic and structural insights. Nat Rev Microbiol. 2014;12:673–685.
- Szemiel AM, Failloux AB, Elliott RM. Role of Bunyamwera Orthobunyavirus NSs protein in infection of mosquito cells. PLoS Negl Trop Dis. 2012;6:e1823.
- Kading RC, Crabtree MB, Bird BH, et al. Deletion of the NSm virulence gene of Rift Valley Fever virus inhibits virus replication in and dissemination from the midgut of Aedes aegypti mosquitoes. PLoS Negl Trop Dis. 2014;8:17–18.
- Won S, Ikegami T, Peters CJ, et al. NSm protein of Rift Valley Fever virus suppresses virus-induced apoptosis. J Virol. 2007;81:13335–13345.
- Sundin DR, Beaty BJ, Nathanson N, et al. A G1 glycoprotein epitope of La Crosse virus: a determinant of infection of Aedes triseriatus. Science. 1987;80:235.
- Plassmeyer ML, Soldan SS, Stachelek KM, et al. California serogroup Gc (G1) glycoprotein is the principal determinant of pH-dependent cell fusion and entry. Virology. 2005;338:121–132.
- Pekosz A, Griot C, Nathanson N, et al. Tropism of bunyaviruses: evidence for a G1 glycoprotein-mediated entry pathway common to the California serogroup. Virology. 1995;214:339–348.
- Hacker JK, Hardy JL. Adsorptive endocytosis of california encephalitis virus into mosquito and mammalian cells: A role for G1. Virology. 1997;235:40–47.
- Hacker JK, Volkman LE, Hardy JL. Requirement for the G1 protein of California encephalitis virus in infection in vitro and in vivo. Virology. 1995;206:945–953.
