ABSTRACT
Bacteriophages are the most abundant organisms on Earth. As there are few effective treatment options against some pathogens, the interest in the bacteriophage control of multi-drug-resistant bacterial pathogens is escalating, especially for Klebsiella pneumoniae. This study aimed to develop a phage-based solution to the rising incidence of extensively drug-resistant clinical Klebsiella pneumoniae sequence type (ST16) infections starting from a set of phages recently characterized against this lineage. A phage-cocktail (Katrice-16) composed of eight lytic phages was characterized for potential use in humans. In vitro and in vivo broth inhibition and Galleria mellonella rescue assays were used to demonstrate the efficacy of this approach using a collection of 56 strains of K. pneumoniae ST16, with distinct genetic backgrounds that were collected from clinical infections from four disparate nations. Additionally, Katrice-16 anti-biofilm activity, synergism with meropenem, and activity in human body fluids were also assessed. Katrice-16 was highly active in vitro against our K. pneumoniae ST16 collection (AUC% median = 86.48%; Q1 = 83.8%; Q2 = 96.85%; Q3 = 98.85%). It additionally demonstrated excellent in vivo activity in G. mellonella rescue assays, even with larvae infected by isolates that exhibited moderate in vitro inhibition. We measured significant anti-biofilm activity over 12 h (p = .0113) and synergic activity with meropenem. In addition, we also demonstrate that Katrice-16 maintained high activity in human body fluids. Our results indicate that our cocktail will likely be an effective solution for human infections with this increasingly prevalent and often highly resistant bacterial clone.
Introduction
The World Health Organization has stated that the expansion of K. pneumoniae resistant to carbapenems and third-generation cephalosporins is of great concern and is a high priority for the development of new antimicrobial agents and novel therapeutic approaches [Citation1]. Several lineages of antibiotic-resistant K. pneumoniae (e.g. ST11, ST15, ST16, ST258, and ST340) are geographically widespread and represent a serious public health issue [Citation2,Citation3]. K. pneumoniae ST16 is an emergent extremely drug resistant (XDR) lineage that has been reported in several countries and is a particular concern in Brazil, where the expansion of this lineage is causing a large burden of human infection [Citation4–9]. Specifically, the common occurrence of horizontally acquired resistance mechanisms such as KPC and NDM together with chromosomal mgrB inactivation that produces colistin resistance in this lineage severely limits therapeutic options and leads to poor prognosis [Citation9].
The limited therapeutic options are currently driving the development of several alternative therapies, including bacteriophages, blue light, nanoparticles, or small peptides [Citation10]. Bacteriophages, viruses that infect and lyse bacteria cells, represent a particularly promising tool. These viruses were initially discovered early in the twentieth century, but their clinical capacity has generally been overlooked in the Western world. However, their utility has increased with the emergence of antimicrobial resistance being especially linked to specific lineages [Citation11,Citation12].
Herein, we developed and tested the in vitro and in vivo efficacy of a bacteriophage cocktail with important antibacterial and anti-biofilm properties against K. pneumoniae ST16.
Material and methods
General features of the bacterial collection
A collection of 56 Klebsiella pneumoniae clinical isolates carrying multiple antimicrobial resistance genes (Table S1) and belonging to ST16 was assembled: 29 previously described in recent studies from Brazil (n = 19) [Citation9] and Italy (n = 10) [Citation8] together with 27 novel isolations from Brazil (n = 25), Bangladesh (n = 1), and Vietnam (n = 1). Susceptibility profiles were determined by disk-diffusion experiments with 16 antibiotics using EUCAST guidelines (Table S1). Broth microdilution and agar dilution were performed to determine the colistin and fosfomycin susceptibility, respectively [Citation13]. The genetic relationship among these isolates was determined by Pulsed Field Gel Electrophoresis after macro-restriction using SpeI-endonuclease and interpreted by the Tenover criteria [Citation14,Citation15].
Bacteriophage cocktail preparation and in vitro killing experiments
Bacteriophages were previously characterized for their genomic and microbiological properties [Citation16], and equal volumes of eight phages (PWKp1, PWKp3, PWKp5, PWKp7, PWKp9B, PWKp14, PWKp15, and PWKp17) at 1 ∼ 3 × 109 PFU/mL, were mixed resulting in a cocktail, Katrice-16, and stored at 4 °C. The general features of each phage present in Katrice-16 are detailed in Table S2.
In order to evaluate the in vitro lytic ability of the cocktail against 56 K. pneumoniae ST16, inhibition experiments were performed: Briefly, 180 μL of an OD ∼ 0.1 (107 K. pneumoniae cells) culture was transferred to 96-well flat plates and mixed with 20 μL of Katrice-16 at 109 PFU/mL resulting in a final MOI of 10. Control samples were added for each isolate (incubation of bacteria plus 20 μL of SM Buffer) for comparative purposes. Optical density (OD600) was measured every 30 min for 12 h with shaking at 200 rpm using a FLUOstar Omega microplate reader (BMG LABTECH Ltd., Aylesbury, UK). Experiments were conducted in triplicates, and inhibition curves were prepared using the average of triplicates with GraphPad Prism (v.8.4.3).
For those isolates poorly inhibited by Katrice-16 MOI 10, we also evaluated different phage/bacteria concentrations on activity. These experiments were conducted as described above, with minor alterations in the bacterial inoculum (start concentration at 106 CFU/mL), followed by increasing Katrice-16 concentration to reach MOIs 10, 100, and 1000 over 16 h of incubation.
Synergism analysis between Katrice-16 and meropenem
Three representative isolates (KL29, OXA-232-producer; BKBR, NDM-1-producer; P28, KPC-2-producer) were selected based on their high level of carbapenem resistance and their inhibition by Katrice-16 MOI 10. Synergy experiments were performed as described by Liu and colleagues [Citation17] with minor modifications to adjust the methodology to the antibiotic susceptibility test recommended by EUCAST/CLSI. 96-well plates were made by combination of 50 μL different meropenem concentrations (0.25–128 mg/L) and 50 μL of Katrice-16 (103–109 PFU/mL) both diluted on cation-adjusted Mueller-Hinton Broth and inoculated to reach a final bacterial concentration of 5 × 105 CFU/mL. The plates were incubated at 37 °C under agitation (200 rpm) and the OD600 was measured every 30 min for 20 h on a FLUOstar Omega microplate reader. The experiments were performed in biological triplicates, and the reduction of bacterial growth was assessed by a mean of these triplicates. Synograms were generated at two-point times (10 and 20 h) based on the deduction of treated wells from the positive control (no treatment) to yield percent reduction: Reduction (%) [(ODgrowthcontrol − ODtreatment)/ODgrowthcontrol] × 100 [Citation17].
Evaluation of in vivo efficacy of Katrice-16 using Galleria mellonella assay
The efficacy of the phage cocktail to rescue fatal infection was evaluated by Galleria mellonella assays as previously described [Citation18] using six K. pneumoniae ST16 strains. We divided the larvae into five major groups: (10 larvae weighing 200–250 mg per each group) based on inoculation in each experiment: (i) control group 1, 10 μL of SM Buffer or 1× PBS; (ii) control group 2, 10 μL of Katrice-16; (iii) infection group, 5 μL of bacteria suspension (106 CFU/mL) + 5 μL of SM Buffer; (iv) treatment group 1, 5 μL of bacteria suspension (106 CFU/mL) + 5 μL of Katrice-16 (107 PFU/mL); (v) treatment group 2, 5 μL of bacteria suspension (106 CFU/mL) + 5 μL of Katrice-16 (108 PFU/mL). All experiments were performed in triplicates.
The bacterial isolates were grown in LB broth for 3 h, centrifuged at 12,000g for 2 min, and washed three times with 1× PBS. Pellets were resuspended in 1× PBS, adjusted to the OD600 = 0.25 ± 0.01, and diluted to reach a final concentration of 106 CFU/mL. Five microliters of bacteria suspension (∼ 5 × 103 CFU) was injected into the last left-side proleg of the larvae using a 10 μL Hamilton syringe (701RN; Hamilton Bonaduz AG), and 5 μL of Katrice-16 (∼ 5 × 104 and 105 PFU), was injected into the last right-side proleg at one-hour post-infection. After treatment, larvae were incubated in the dark at 37 °C and their survival status checked at 24, 48, and 72 h.
Katrice-16 anti-biofilm activity
Katrice-16 activity against mature biofilms produced by 30 randomly selected K. pneumoniae ST16 isolates was assessed. Biofilms were grown on Tryptic soy broth (TSB) supplemented with 1% glucose for 48 h with change of bacterial media after 24 h. After two days of biofilm growth, two groups were created: (i) an untreated group, that received 200 μL of TSB broth with 1% glucose; (ii) and a treated group, which received 108 PFU/mL of Katrice-16 diluted in 200 μL of TSB broth with 1% glucose. We conducted the treatment over 12 and 24 h; After treatment, the plates were washed with PBS 1X, stained with crystal violet (0.2%) for 30 min and eluted in 30% (vol/vol) glacial acetic acid, as previously described [Citation18]. The absorbance of each well was measured at 595 nm and each experiment was conducted in triplicate.
Bacteriophage cocktail activity in human body fluids
Urine from five (male n = 4 and female n = 1) healthy donors was collected, warmed to 37 °C and tested against bacteria (uroculture) with pH verification. The samples were pooled, filtered through a 0.22 μM membrane and stored at −20 °C. Prior to each test, urine samples were defrosted and kept for 4 h at 37 °C with agitation every hour. To simulate a urinary tract infection, where the pH is generally more acidic, we used HCl 1M to adjust the pH of the second urine sample. Two urine pools were tested: (i) the original urine pool, pH 6.5 and (ii) a urine pool at pH 5.5. Katrice-16 was diluted in the urine samples, reaching a final concentration of ∼5 × 108 PFU/mL and its activity was checked by comparing the bacterial growth in the presence/absence of phage cocktail. For urine inhibition tests, 180 μL of Katrice-16 diluted in urine was transferred into 96-well flat plates and mixed with 20 μL of bacterial inoculum in LB broth, resulting in a final OD600 of ∼0.1 (∼107 CFU/mL). Thus, all experiments were carried out having 10% LB broth. Control samples were added for each isolate (incubation of 180 μL urine sample, without bacteriophage plus 20 μL of bacteria inoculum) for comparative purposes. Optical density (OD600) was measured every 30 min interval over 12 h with shaking at 200 rpm using FLUOstar Omega microplate reader (BMG LABTECH Ltd., Aylesbury, UK). These experiments were performed in triplicate, and the results were analysed and interpreted using GraphPad Prism (v.8.4.3).
Pooled, pre-inactivated human AB serum was used to check Katrice-16 stability and activity against the same isolates used in the urine analysis. Once thawed, the serum was kept for 1 h at 37 °C and then mixed with Katrice-16 at a final concentration of ∼5 × 108 PFU/mL. For inhibition tests, 180 μL of Katrice-16 diluted in human serum was transferred into 96-well flat plates and mixed with 20 μL of bacterial inoculum in LB broth, resulting in a final OD600 of ∼0.1 (∼107 CFU/mL). Control samples were added for each isolate (incubation of 180 μL human serum, without bacteriophage plus 20 μL of bacteria inoculum) for comparative purposes. The measurement of results and interpretation were performed as described above.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (version 8.4.3, San Diego, CA, USA). One-way ANOVA followed by multiple comparison tests were used to compare AUC% and Katrice-16 activity in different human body fluids. Multiple comparison tests applied in each analysis are described in their respective figure legends. Differences in biofilm removal were evaluated by Two-tailed t tests. Log-rank (Mantel–Cox) test was used for survival curves rescue experiments. P-values of <.05 were considered statically significant.
Results
In vitro activity of Katrice-16 against XDR K. pneumoniae ST16 collection
Katrice-16 demonstrated excellent in vitro activity against the K. pneumoniae ST16 collection, inhibiting bacterial growth of 98% of bacterial isolates from 12 h ((A)). The distribution of all AUCs% ((B)) was high, indicating effective lysis (AUC% median = 86.48%; Q1 = 83.8%; Q2 = 96.85%; Q3 = 98.85%), especially for those isolates recovered from Brazil (AUC% median = 89.62) and Italy (AUC% median = 80.39). The two Asian isolates evaluated had a non-satisfactory inhibition (BKBR, AUC% = 29.5) and a moderate inhibition (VNV8, AUC% = 66.2) ((A); Figure S1).
Figure 1. Inhibition activity of Katrice-16 against K. pneumoniae ST16. (A) Representative broth inhibition assays of Katrice-16 MOI 10 and K. penumoniae ST16 over 12 h, error bars represent the SD (n = 3). For each graph, the area under curve (AUC%) was calculated based on the bacterial growth in the presence/absence of Katrice-16 MOI 10. AUC% below 50% are highlighted in red. The set with the inhibition curves of all K. pneumoniae ST16 isolates can been seen in Figure S1. (B) Violin plot demonstrating the distribution of all 56 K. pneumoniae ST16 AUC% using Katrice-16 MOI 10. The blue squares are individual values of AUC%, the red line represents the mean, and the blue lines the quartiles (Q1, Q2, and Q3). (C) Broth inhibition assays of representative isolates poorly inhibited by Katrice-16 MOI 10. During this new assay, an increased concentration of Katrice-16 was used to achieve higher MOIs (100 and 1000). The experiment was conducted over 16 h and error bars represent the SD (n = 3). D Individual AUC% values of the eight poorly inhibited isolates submitted to a new broth inhibition test using higher MOIs. Black lines represent the median for each group and p value was calculated by one-way analysis of variance (ANOVA) followed by Tukey multiple comparison post hoc test [MOI 10 vs. MOI 100, p = .1185; MOI 10 vs. MOI 1000, p = .0164; MOI 100 vs. MOI 1000, p = .6085].
![Figure 1. Inhibition activity of Katrice-16 against K. pneumoniae ST16. (A) Representative broth inhibition assays of Katrice-16 MOI 10 and K. penumoniae ST16 over 12 h, error bars represent the SD (n = 3). For each graph, the area under curve (AUC%) was calculated based on the bacterial growth in the presence/absence of Katrice-16 MOI 10. AUC% below 50% are highlighted in red. The set with the inhibition curves of all K. pneumoniae ST16 isolates can been seen in Figure S1. (B) Violin plot demonstrating the distribution of all 56 K. pneumoniae ST16 AUC% using Katrice-16 MOI 10. The blue squares are individual values of AUC%, the red line represents the mean, and the blue lines the quartiles (Q1, Q2, and Q3). (C) Broth inhibition assays of representative isolates poorly inhibited by Katrice-16 MOI 10. During this new assay, an increased concentration of Katrice-16 was used to achieve higher MOIs (100 and 1000). The experiment was conducted over 16 h and error bars represent the SD (n = 3). D Individual AUC% values of the eight poorly inhibited isolates submitted to a new broth inhibition test using higher MOIs. Black lines represent the median for each group and p value was calculated by one-way analysis of variance (ANOVA) followed by Tukey multiple comparison post hoc test [MOI 10 vs. MOI 100, p = .1185; MOI 10 vs. MOI 1000, p = .0164; MOI 100 vs. MOI 1000, p = .6085].](/cms/asset/21ca2452-52c5-4ed4-8793-4b7c8f0afc2f/temi_a_2051752_f0001_oc.jpg)
We also noticed that increasing the concentration of Katrice-16 could dramatically improve the cocktail inhibition. All strains demonstrated an increase of inhibition as higher MOI were used (MOI 100 and MOI 1000) with the sole exception of isolate P32. For some isolates (e.g. P28, P31, and KL1), this increase resulted in a complete inhibition of bacterial growth, demonstrating that the relationship between the number of phages per bacterium can be a critical point for the outcome of the inhibition of bacterial growth ((C)). Tukey’s test revealed a significant difference between tests using MOI 10 and MOI 1000 (p = .0164) ((D)).
In vivo Katrice-16 therapy rescues G. melonella from infection
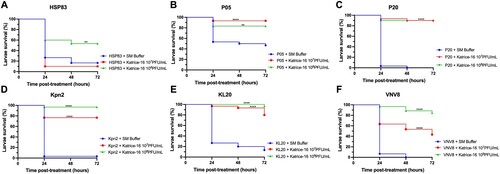
Using a single Katrice-16 cocktail injection (1-hour after infection) as treatment had a dramatic impact on larvae survival for all evaluated isolates (). Larvae survival rates after treatment with 107 and 108 PFU/mL of Katrice-16 increased from 0% to 90% (p < .0001 for both cocktail concentrations) for larvae infected with P20. This was also seen for Kpn2, increasing survival rates from 3.3% to 76.6% (p < .0001, 107 PFU/mL) and 96.6% (p < .0001, 108 PFU/mL) (). The concentration of Katrice-16 was also important for some experimental outcomes. Whist no or a slight difference was observed between the two cocktail concentrations for P05, P20, and KL20, the use of a higher concentration of Katrice-16 resulted in better outcomes for the remaining strains (). The controls used in this study indicated that Katrice-16, PBS or SM buffer had no adverse effects on the larvae (Figure S2).
Figure 2. In vivo efficacy of Katrice-16 demonstrated by Kaplan–Meier plots showing the percent of G. mellonella survival after phage cocktail treatment. P values were calculated using Log-rank (Mantel-Cox) test for (A) (107 PFU/mL, p = .3114, χ² = 1.025, d.f = 1; 10⁸ PFU/mL, p = .0027, χ² = 8.997, d.f = 1), (B) (10⁷ PFU/mL, p < .0001, χ² = 15.43, d.f = 1; 10⁸ PFU/mL, p = .0032, χ² = 8.716, d.f = 1), (C) (10⁷ PFU/mL, p < .0001, χ² = 53.76, d.f = 1; 10⁸ PFU/mL, p < .0001, χ² = 50.90, d.f = 1), (D) (10⁷ PFU/mL, p < .0001, χ² = 33.05, d.f = 1; 10⁸ PFU/mL, p < .0001, χ² = 51.40, d.f = 1), (E) (10⁷ PFU/mL, p < .0001, χ² = 33.91, d.f = 1; 10⁸ PFU/mL, p < .0001, χ² = 46.54, d.f = 1), (F) (10⁷ PFU/mL, p < .0001, χ² = 26.34, d.f = 1; 10⁸ PFU/mL, p < .0001, χ² = 56.86, d.f = 1). *indicates .01 < p value < .05, **indicates .001 < p value < .01, ***indicates .0001 < p value < .001, ****indicates p value < .0001.
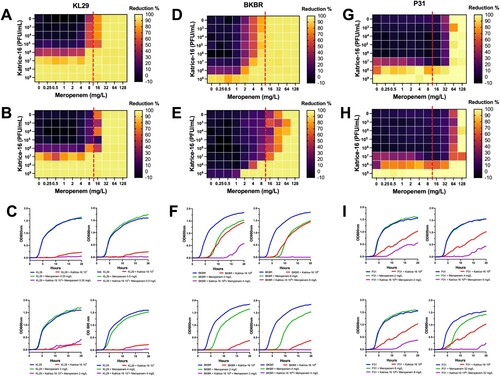
Katrice-16 impact on bacterial growth when associated with meropenem and its anti-biofilm activity
The OXA-232-producing K. pneumoniae ST16 (KL29) showed an initial meropenem MIC of 32 mg/L and did not show any synergism when sub-inhibitory concentrations of meropenem were combined with Katrice-16 ranging from 103–106 PFU/mL (). However, a significant reduction in bacterial growth was observed when 107 PFU/mL was combined with sub-inhibitory concentrations of meropenem (0.25–8 mg/L), reducing the original MIC to 8 mg/mL. Synergistic results were also obtained for NDM-1-producing K. pneumoniae ST16 (BKBR), dropping the original MIC from 64 to 2 mg/L when associated with 109 PFU/mL of Katrice-16 (). Among the three tested isolates, BKBR showed the highest variation between 10 and 20 h of analysis and the combination of sub-inhibitory concentrations of meropenem (2, 4, and 8 mg/L) where 109 PFU/mL. P31 synergism resulted in a subtle drop of meropenem MIC (128–64 mg/mL) when combined with 108 PFU/mL of Katrice-16. Although less significant than the BKBR synergism, the association of sub-inhibitory concentrations and 108 PFU/mL of Katrice-16 resulted in a reduction of bacterial growth by 70–80% for 2, 4, and 32 mg/mL of meropenem, demonstrating the potential clinical impact of combination therapy.
Figure 3. Synergism activity between Katrice-16 and meropenem against three carbapenemase-producing K. pneumoniae ST16 isolates. Synograms were constructed based on the data of bacterial growth in the presence/absence of antibiotic and phages over 10 and 20 h. Dashed red lines represent the susceptibility breakpoint of meropenem according to EUCAST, 2021. KL29 synograms from 10 h (A), and 20 h (B) followed by a set of four selected growth curves (C), demonstrating the effect of different concentrations of meropenem and Katrice-16 in the bacteria growth. BKBR synograms from 10 h (D), and 20 h (E) followed by a set of four selected growth curves (F), demonstrating the effect of different concentrations of meropenem and Katrice-16 in the bacteria growth. P31 synograms from 10 h (G), and 20 h (H) followed by a set of four selected growth curves (I), demonstrating the effect of different concentrations of meropenem and Katrice-16 in the bacterial growth.
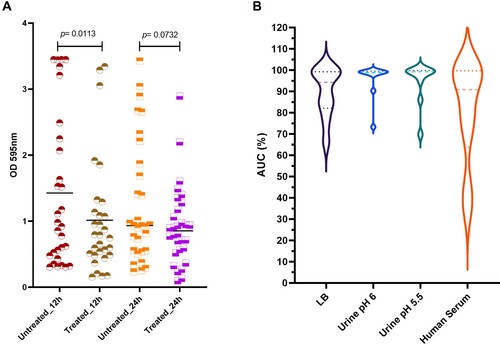
Katrice-16 was also able to combat the mature biofilm produced by K. pneumoniae ST16 isolates ((A)). The median biofilm production of the 12 h-treated group was lower than 12 h-untreated group using the cocktail at 108 PFU/mL, suggesting an efficient biofilm reduction (Wilcoxon test, p = .0113). Interestingly, the results were more promising for those isolates with higher biofilm production. In the untreated group, nine isolates had values of OD595nm higher than 2, while after 12-hour treatment, only three isolates remained with values above 2. On the other hand, although the average of the 24 h-treated group was lower than 24 h-untreated group, no statistical difference was observed between the groups (Wilcoxon test, p = .0732).
Figure 4. Katrice-16 antibiofilm activity and its performance in human body fluids. (A) Biofilm production of 30 K. pneumoniae ST16 isolates with and without treatment (12 and 24 h). Black horizontal lines represent the median of each group and p value was calculated by Wilcoxon signed-ranks test (12 h, p = .0113; 24 h, p = .0732). (B) Katrice-16 activity demonstrated by AUC% values of 10 selected K. pneumoniae ST16 in Lysogeny Broth (LB) and human body fluids (urine and human serum). Violin plots’ internal lines represent median and quartiles, and p values were calculated using non-parametric one-way ANOVA (Tukey’s multiple comparisons tests).
Activity of Katrice-16 in different human body fluids
Katrice-16 lytic activity was also evaluated in urine and human serum. Urine inhibition assays demonstrated good activity of Katrice-16, with no statistical difference in the results of inhibition obtained with both urine samples at pH 6.5 (p = .8052) and at pH 5.5 (p = 0.8676) as compared to tests performed in LB broth by Tukey's multiple comparison test. The reduction of urine pH did not have any effect on Katrice-16 activity since results of AUC% were identical (p = .9992). Effective Katrice-16 activity was also conserved in human serum, with no significant difference when compared to LB broth (p = 0.5483). There was a small AUC% median decrease in comparison to urine tests (AUC% = 80.71 of human serum vs. AUC% = 95.63 and 94.76 of urine pH 5.5 and pH 6.5, respectively). However, this difference was not statistically relevant, as observed by Tukey’s test ((B)).
Discussion
Katrice-16 is a phage cocktail approach developed to combat K. pneumoniae ST16 isolates of different genetic backgrounds. The cocktail was designed by mixing eight Klebsiella phages belonging to three different phage families. Although experiments to characterize the infection pathway of each bacteriophage are underway, it is likely that the different families of bacteriophages target different bacterial receptors and as such limit the emergence of phage-resistant mutants. Due to host-range results, phages belonging to the same family were also added to the cocktail. We tested different phage combinations to significantly create the best cocktail able to inhibit our tested collection. Our cocktail demonstrated excellent inhibition of the growth of the majority of XDR K. pneumoniae ST16 strains isolated from several disparate countries. This was demonstrated by both in vitro growth inhibition assays and by in vivo tests using the G. mellonella model ((B) and ). Although some isolates were genetically related by PFGE (Table S1), they varied in phage susceptibility (Figure S1). Point mutations on phage receptors, alteration of receptor expression or capsule production can modify the phage susceptibility exhibited by a strain [Citation19,Citation20]. Thus, we suggested that we should not only rely on the PFGE clonal relationship of different strains to predict phage susceptibility. The rapid emergence and dissemination of antimicrobial resistance determinants have led to difficulties in the treatment of human infection and calls for alternative approaches. This is especially important since antibiotic use itself is a driver of both emergence and dissemination of resistance [Citation21]. Successful in vitro and in vivo use of bacteriophages has been widely demonstrated in recent years [Citation22–24], and detailed studies such as this one are necessary to ensure the robustness of bacteriophage use. Our study is important as it demonstrates that it is possible to develop a general cocktail capable of efficiently combating different isolates belonging to the same lineage from different countries. This is of major importance since it is now recognized that antibiotic resistance mechanisms are very often associated with a small number of prevalent STs of each species [Citation25].
In vivo experiments using the G. mellonella assay also demonstrated the efficacy and safety of our cocktail. Our experiments establish that the cocktail has an impressive ability to rescue fatal infections. This is especially demonstrated for those isolates with lower AUC% values such as KL20 and VNV8 (). This data is also important because it reveals that an excellent in vitro inhibition is not an absolute requirement for a satisfactory in vivo activity. Recent studies have also demonstrated the therapeutic efficacy of bacteriophage cocktails against several clinically relevant micro-organisms including K. pneumoniae, Pseudomonas aeruginosa and A. baumannii and demonstrates the utility and reproducibility of using bacteriophages to rescue organisms infected by these pathogens [Citation18,Citation26,Citation27].
We tested the hypothesis that meropenem could be used in combination with Katrice-16, even for carbapenem-resistant bacteria, establishing that the combination could be useful to combat K. pneumoniae ST16. This finding represents a promising alternative to more efficiently treat bacterial isolates resistant to carbapenems. Similar results have been obtained combining meropenem (16–256 mg/L) with low concentrations of the myovirus KARL-1 (MOI 10−1–10−7), with significantly decreased MDR A. baumannii growth, demonstrating that our finding can also be applied to other gram negatives [Citation28].
Katrice-16 additionally exhibited good anti-biofilm activity, especially at 12-h post-treatment ((A)). We believe that the superior results obtained at 12-h versus 24-h treatment are related to the age of the biofilm since it is known that phages have higher efficiency in infecting cells with high metabolic activity, exactly the opposite of what is found in biofilms [Citation29,Citation30]. Although we only tested Katrice-16 anti-biofilm activity at 37 °C, we believe that at RT, the cocktail should remain effective since the bacteriophages demonstrated good activity at RT [Citation25]. This is important in the case of K. pneumoniae ST16 since this microorganism has a great capacity to persist in hospital settings and on abiotic surfaces and as such, cause complex prosthetic joint infections [Citation31].
Conclusion
As new reports emerge indicating the spread of MDR K. pneumoniae ST16 [Citation5,Citation32], we have demonstrated that phage-based approaches are an attractive way to combat these micro-organisms due to their low cost, their good in vitro and in vivo activity and to their effectiveness in human body fluids. The rapidity of this approach is in stark contrast to the time taken to discover and evaluate an effective new antibiotic, and there is therefore great potential in using this approach to counteract emerging MDR pathogens such as K. pneumoniae ST16.
Supplemental Material
Download Zip (740.7 KB)Acknowledgements
The authors would like to thank Diego Andrey and Juliana Paulino for their efforts in the initial characterization of K. pneumoniae ST16 isolates, Kesia Silva and Miriam R. Fernandes for their suggestions for this study and Brad Spiller for his contribution to human serum experiments. W. M. B. S. M., A. C. G., M. T. conceived and designed the experiments; W. M. B. S. M., M. L., K. S., E. P., J. M. performed the experiments; W. M. B. S. M., K. S., B. H., P. D., R. M., E. A. M., A. C. G., M. T. provided sewage samples and bacterial strains; W. M. B. S. M., J. R. H., M. T. analysed the data; A. C. G., M. T. secured funds and provided resources; W. M. B. S. M. helped in writing of the original manuscript; and K. S., E. P., and M. T. contributed to the edition of the manuscript. All authors contributed extensively to work presented in this paper. All authors discussed the results and implications and commented on the manuscript at all stages.
Disclosure statement
A. C. G. has recently received research funding and/or consultation fees from Cristália, Enthasis Therapeutics, InfectoPharm, Eurofarma, Pfizer, MSD, and Zambon. Other authors have nothing to declare.
Additional information
Funding
References
- Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327.
- Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. 2015;112(27):E3574–E3581.
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275.
- Lester CH, Olsen SS, Jakobsen L, et al. Emergence of extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae in Danish hospitals; this is in part explained by spread of two CTX-M-15 clones with multilocus sequence types 15 and 16 in Zealand. Int J Antimicrob Agents. 2011;38(2):180–182.
- Boonyasiri A, Jauneikaite E, Brinkac LM, et al. Genomic and clinical characterisation of multidrug-resistant carbapenemase-producing ST231 and ST16 Klebsiella pneumoniae isolates colonising patients at Siriraj hospital, Bangkok, Thailand from 2015 to 2017. BMC Infect Dis. 2021;21(1):142.
- Marcade G, Brisse S, Bialek S, et al. The emergence of multidrug-resistant Klebsiella pneumoniae of international clones ST13, ST16, ST35, ST48 and ST101 in a teaching hospital in the Paris region. Epidemiol Infect. 2013;141(8):1705–1712.
- Moradigaravand D, Martin V, Peacock SJ, et al. Evolution and Epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio. 2017;8(1):e01976–16.
- Espinal P, Nucleo E, Caltagirone M, et al. Genomics of Klebsiella pneumoniae ST16 producing NDM-1, CTX-M-15, and OXA-232. Clin Microbiol Infect. 2019;25(3):385.e1–385.e5.
- Andrey DO, Pereira Dantas P, Martins WMBS, et al. An Emerging clone, Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae sequence type 16, associated with high mortality rates in a CC258-endemic setting. Clin Infect Dis. 2020;71(7):e141–e150.
- Roca I, Akova M, Baquero F, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015;6:22–29. Erratum in: New Microbes New Infect 2015. 8. 175.
- Salmond GP, Fineran PC. A century of the phage: past, present and future. Nat Rev Microbiol. 2015;13(12):777–786.
- Martins WMBS, Toleman MA, Gales AC. Clinical utilization of bacteriophages: a new perspective to combat the antimicrobial resistance in Brazil. Braz J Infect Dis. 2020;24(3):239–246.
- European Committee on Antimicrobial Susceptibility Testing. Clinical breakpoints and dosing of antibiotics. [cited 2021 May 10]. Available from: http://www.eucast.org/clinical_breakpoints/.
- Centers for Disease Control and Prevention. PulseNet standard protocol. [cited 2021 Feb 20]. Available at: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfgeprotocol-508c.pdf.
- Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239.
- Martins WMBS, Cino J, Li M, et al. Lytic bacteriophages against mutidrug-resistant Klebsiella pneumoniae: development of an effective phage-based approach to combat multidrug resistance. Research Square. 2021. Sep 07; doi: 10.21203/rs.3.rs-850585/v1.
- Gu Liu C, Green SI, Min L, et al. Phage-antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. MBio. 2020;11(4):e01462–20.
- Jeon J, Yong D. Two novel bacteriophages improve survival in Galleria mellonella infection and mouse acute pneumonia models infected with extensively drug-resistant Pseudomonas aeruginosa. Appl Environ Microbiol. 2019;85(9):e02900–18.
- Wright RCT, Friman VP, Smith MCM, et al. Resistance evolution against phage combinations depends on the timing and order of exposure. mBio. 2019;10(5):e01652–19.
- Fang Q, Feng Y, McNally A, et al. Characterization of phage resistance and phages capable of intestinal decolonization of carbapenem-resistant Klebsiella pneumoniae in mice. Commun Biol. 2022;5(1):48.
- Kumar M, Sarma DK, Shubham S, et al. Futuristic non-antibiotic therapies to combat antibiotic resistance: A review. Front Microbiol. 2021;12:609459.
- Zurabov F, Zhilenkov E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol J. 2021;18(1):9.
- Corbellino M, Kieffer N, Kutateladze M, et al. Eradication of a multidrug-resistant, carbapenemase-producing Klebsiella pneumoniae isolate following oral and intra-rectal therapy With a custom made, Lytic Bacteriophage preparation. Clin Infect Dis. 2020;70(9):1998–2001.
- Manohar P, Tamhankar AJ, Lundborg CS, et al. Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and enterobacter species. Front Microbiol. 2019;10:574.
- Martins W, Mathias J, Babenko D, et al. Engineering the human microbiome, A natural double-barreled approach towards solving both our current antibiotic resistance and sepsis problems. Am J Biomed Sci Res. 2020;11(3). AJBSR.MS.ID.001632.
- Jeon J, Park JH, Yong D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019;19(1):70.
- Thiry D, Passet V, Danis-Wlodarczyk K, et al. New bacteriophages against emerging lineages ST23 and ST258 of Klebsiella pneumoniae and efficacy assessment in Galleria mellonella larvae. Viruses. 2019;11(5):411.
- Jansen M, Wahida A, Latz S, et al. Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multi-drug resistant Acinetobacter baumannii. Sci Rep. 2018;8(1):14140.
- Azeredo J, Sutherland IW. The use of phages for the removal of infectious biofilms. Curr Pharm Biotechnol. 2008;9(4):261–266.
- Abedon ST. Ecology of anti-biofilm agents II: bacteriophage exploitation and biocontrol of biofilm bacteria. Pharmaceuticals (Basel). 2015;8(3):559–589.
- de Sanctis J, Teixeira L, van Duin D, et al. Complex prosthetic joint infections due to carbapenemase-producing Klebsiella pneumoniae: a unique challenge in the era of untreatable infections. Int J Infect Dis. 2014;25:73–78.
- Nguyen TNT, Nguyen PLN, Le NTQ, et al. Emerging carbapenem-resistant Klebsiella pneumoniae sequence type 16 causing multiple outbreaks in a tertiary hospital in southern Vietnam. Microb Genom. 2021;7(3):mgen000519.
