ABSTRACT
There is a limited understanding of the pathophysiology of postacute pulmonary sequelae in severe COVID-19. The aim of current study was to define the circulating microRNA (miRNA) profiles associated with pulmonary function and radiologic features in survivors of SARS-CoV-2-induced ARDS. The study included patients who developed ARDS secondary to SARS-CoV-2 infection (n = 167) and a group of infected patients who did not develop ARDS (n = 33). Patients were evaluated 3 months after hospital discharge. The follow-up included a complete pulmonary evaluation and chest computed tomography. Plasma miRNA profiling was performed using RT-qPCR. Random forest was used to construct miRNA signatures associated with lung diffusing capacity for carbon monoxide (DLCO) and total severity score (TSS). Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses were conducted. DLCO < 80% predicted was observed in 81.8% of the patients. TSS showed a median [P25;P75] of 5 [2;8]. The miRNA model associated with DLCO comprised miR-17-5p, miR-27a-3p, miR-126-3p, miR-146a-5p and miR-495-3p. Concerning radiologic features, a miRNA signature composed by miR-9-5p, miR-21-5p, miR-24-3p and miR-221-3p correlated with TSS values. These associations were not observed in the non-ARDS group. KEGG pathway and GO enrichment analyses provided evidence of molecular mechanisms related not only to profibrotic or anti-inflammatory states but also to cell death, immune response, hypoxia, vascularization, coagulation and viral infection. In conclusion, diffusing capacity and radiological features in survivors from SARS-CoV-2-induced ARDS are associated with specific miRNA profiles. These findings provide novel insights into the possible molecular pathways underlying the pathogenesis of pulmonary sequelae.
Trial registration: ClinicalTrials.gov identifier: NCT04457505..
Trial registration: ISRCTN.org identifier: ISRCTN16865246..
Introduction
Persistent pulmonary dysfunction has been described as a common clinical feature in survivors of acute respiratory distress syndrome (ARDS) [Citation1,Citation2]. Indeed, 15-50% of patients who recovered from ARDS as a consequence of coronavirus pneumonia, such as in severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), experienced major long-term sequelae after hospital discharge (3-24 months), including impaired lung diffusing capacity for carbon monoxide (DLCO) and radiological abnormalities [Citation3–5]. Our group recently demonstrated that up to 80% of survivors of ARDS secondary to SARS-CoV-2 infection showed diffusion impairment and pulmonary structural alterations in a 3-month follow-up study [Citation6]. Similar findings have been reported in different cohorts of discharged COVID-19 patients [Citation7–11].
Given the significant burden of pulmonary sequelae caused by SARS-CoV-2 and the current lack of evidence to treat lung dysfunction in “post-COVID” syndrome, the development of innovative therapies is of vital importance. In this scenario, molecular phenotyping of the patient emerges as an essential tool to identify the underlying biological factors implicated in the sequelae and, consequently, targets for intervention [Citation12].
MicroRNAs (miRNAs) are single-stranded noncoding RNA sequences (19-25 nt) that function as posttranscriptional repressors of gene expression. miRNAs participate in complex and coordinated regulatory networks [Citation13], with a single miRNA regulating large sets of genes and a single gene able to be targeted by large sets of miRNAs. This biological system allows the fine-tuning of the cellular phenotype. As such, miRNAs constitute essential modulators of cellular development, homeostasis and response to stress [Citation14]. miRNAs have also been described in the extracellular space [Citation15], including biofluids such as plasma [Citation16]. Extracellular miRNAs could act as signalling molecules at the paracrine and endocrine levels to regulate a broad array of physiological processes [Citation17,Citation18]. Their aberrant expression plays a significant role in the onset and development of pathological conditions, including pulmonary diseases [Citation19,Citation20].
Here, we evaluated the association between pulmonary function and radiologic features and the circulating miRNA profile in survivors of SARS-CoV-2-induced ARDS 3 months after hospital discharge. The final aim is to decipher the pathophysiology of the postacute pulmonary sequelae of severe COVID-19. To the best of our knowledge, this is the first study that uses an approach based on the miRNA transcriptome to characterize the pulmonary function and structural outcomes of ARDS secondary to SARS-CoV-2 infection.
Methods
Ethics statements
The study protocol was approved by the medical ethics committee of Hospital Universitari Arnau de Vilanova (CEIC/2273) and the Institutional Review Board (IRB) from Servicio de Salud Bio (IRB: CEC113) and Servicio de Salud Concepcion (IRB: CEC-SSC: 20-07-26). The study protocol was registered in the ISRCTN registry (ID: ISRCTN16865246). The study was performed in full compliance with the Declaration of Helsinki. The patients received written information about the nature and goals of the study and signed an informed consent form. Patient data and sample data are kept in a database with restricted access.
Study design and population
This is a substudy of the ongoing multicenter study CIBERESUCICOVID registered at www.clinicaltrials.gov with the identification NCT04457505 [Citation21]. This was a prospective study including patients admitted due to severe COVID-19 at Hospital Universitari Arnau de Vilanova-Santa María (Lleida, Spain) between March and December 2020. Patients were evaluated in a “post-COVID” consultation 3 months after hospital discharge in the same hospital (median [P25;P75] of 96.00 [85.75;107.00] days). The final study population was selected according to the following inclusion criteria: i) positive for SARS-CoV-2 according to a standardized test; ii) aged over 18; iii) developed ARDS according to the Berlin definition [Citation22] during hospital stay; and iv) attended a “post-COVID” consultation. The exclusion criteria for the “Post-COVID” evaluation were: i) less than 1 year of life expectancy; ii) transferred to another institution; iii) treated with palliative care; and iv) impossible to evaluate performance due to severe mental disability. A control group composed by patients positive for SARS-CoV-2 but who did not develop ARDS was additionally included. Patients were evaluated at Hospital Universitari Arnau de Vilanova-Santa María (Lleida, Spain), Hospital Complejo Víctor Ríos Ruiz (Los Ángeles, Chile) and Hospital Guillermo Grant Benavente (Concepción, Chile) between July 2020 and August 2021. Demographic, clinical, pharmacological and laboratory data were abstracted from the electronic medical records and entered into a REDCap database.
Pulmonary evaluation
A complete pulmonary evaluation was performed as previously described [Citation6]. Airway function was measured using a flow spirometer (MasterScreen, Jaeger, Germany) according to the guidelines of the American Thoracic Society [Citation23]. The results were expressed as a percentage of the DLCO predicted value according to the European Community Lung Health Survey [Citation24]. Chest computed tomography (CT) examinations were performed using a sixteen- and sixty-four-slice multidetector CT scanner (Brilliance 16 and 64; Philips Healthcare). Images were acquired with patients in the supine position in the cranio-caudal direction at end-inspiration. To quantify the severity of lung affectation, the total severity score (TSS) was assessed. Each of the five lung lobes was determined for the percentage of lobar involvement. Following this, the severity of each lobe was classified as none (0%), minimal (1-25%), mild (26-50%), moderate (51-75%), or severe (76-100%), with a corresponding score of 0, 1, 2, 3 or 4, respectively. The TSS is calculated by summing the five lobe scores (range from 0 to 20) [Citation25].
Samples
Samples collected at the Hospital Universitari Arnau de Vilanova-Santa María (Lleida, Spain) were processed in standardized conditions with support by IRBLleida Biobank (B.0000682) and “Plataforma Biobancos PT20/00021”. Samples collected at the University of Concepción (Concepción, Chile) were processed using standardized conditions according to international Biobank regulations. Venous blood samples were obtained in ethylenediaminetetraacetic acid (EDTA) blood collection tubes (BD, NJ, USA) by venipuncture after a night of fasting and before beginning any interventional procedure at the “post-COVID” consultation 3 months after hospital discharge. To obtain plasma, blood samples were fractionated by centrifugation at 1,500 xg for 10 min at room temperature. After centrifugation, the plasma supernatant was immediately aliquoted, frozen and stored at −80°C until analysis. Samples collected in University of Concepción (Concepción, Chile) were shipped on dry ice to the IRBLleida Biobank.
Antigen detection in plasma
Panbio® COVID-19 Ag Rapid Test Device (Abbott, Chicago, IL, USA) was used to detect the N antigen of SARS-CoV-2 in the plasma samples.
MicroRNA quantification
RNA isolation and miRNA quantification were conducted in the same laboratory using standardized protocols. Experienced staff blinded to patient data performed the miRNA quantification.
Total RNA was isolated from 200 μL of frozen plasma using the miRNeasy Serum/Plasma Advanced kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. For normalization, synthetic Caenorhabditis elegans miR-39-3p (cel-miR-39-3p), lacking sequence homology to human miRNAs, was added as an external reference miRNA (1.6 × 108 copies/μL). The mixture was supplemented with 1 µg of MS2 carrier RNA (Roche Diagnostics, Mannheim, Germany), not containing miRNAs, to improve extracellular miRNA yield. The RNA Spike-In Kit (UniSp2, UniSp4 and UniSp5) (Qiagen, Hilden, Germany) was added to monitor RNA isolation. All reagents were spiked into samples during RNA isolation after incubation with the denaturing solution. RNA was eluted with 20 μL of RNAse-free water and stored at −80°C.
miRNA quantification was performed according to the protocol from the miRCURY LNA Universal RT microRNA PCR System (Qiagen, Hilden, Germany), which offers optimal analytical performance [Citation26]. Reverse transcription (RT) cDNA synthesis was performed using a miRCURY LNA RT Kit (Qiagen, Hilden, Germany) in a total reaction volume of 10 μL. An additional spike-in UniSp6 (Qiagen, Hilden, Germany) was added to monitor the RT reaction. The RT reactions were performed under the following conditions: incubation for 60 min at 42°C, inactivation for 5 min at 95°C, and immediate cooling to 4°C. Then, cDNA was stored at 20°C. A panel of 41 miRNAs (Supplemental Table S1), previously described [Citation27], was analyzed using miRCURY LNA miRNA Custom Panels (384-well plates) (Qiagen, Hilden, Germany). qPCR was carried out using the QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) in a total volume of 10 μL. RT-qPCR conditions were 95°C for 2 min, followed by 40 cycles of 95°C for 10 s and 56°C for 1 min, followed by melting curve analysis. Synthetic UniSp3 was analyzed as an interplate calibrator and qPCR control. Amplification curves were evaluated using QuantStudio Software v1.3 (Thermo Fisher Scientific, Massachusetts, USA).
The quantification cycle (Cq) was defined as the fractional cycle number at which the fluorescence exceeded a given threshold. The specificity of the qPCR was corroborated by melting curve analysis. To ensure the optimal quality of the data, the spike-in RNA templates were first analyzed to monitor the uniformity of the RNA extraction procedure and the efficiency of the RT and PCR. Hemolysis contamination was excluded as previously described [Citation28]. miRNAs were considered to be expressed at Cq values < 35. Cqs above 35 cycles were considered undetectable and were censored at the minimum level observed for each miRNA. Relative quantification was performed using the 2-ΔCq method (ΔCq = CqmiRNA-Cqcel-miR-39-3p). Expression levels were log-transformed for statistical purposes.
Molecular pathway and Gene Ontology analyses
Bioinformatic prediction analysis was performed using the web-based computational tool DIANA-miRPath v3.0 [Citation29]. DIANA-miRPath v3.0 combines information on manually curated experimentally validated miRNA:gene interactions from TaRBase v7.0 with the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and Gene Ontology (GO) annotations (for biological processes). Enrichment analysis was performed using Fisher’s exact test (hypergeometric distribution). The false discovery rate (FDR)-adjusted p-value was set at <0.05. A Venn diagram was generated to represent shared and unique molecular pathways and biological processes for pulmonary function and radiologic features.
Statistical analysis
Descriptive statistics were used to summarize the characteristics of the study population. Data are presented as the median [P25;P75] for continuous variables and as frequencies (percentage) for categorical variables. Due to the high burden of diffusion impairment and the lack of clinical cutoff for radiological features, both DLCO and TSS were analyzed as tertiles and/or continuous variables. To identify a dose–response relationship between individual miRNAs and the outcomes, differential expression was based on p for trend using linear models for arrays [Citation30]. The models included miRNA levels as outcome and study variables categorized by tertiles (transformed as: first tertile = 1; second tertile = 2; third tertile = 3). Differential expression between study groups is displayed in volcano plots. The multivariable analysis was performed using random forest (RF). DLCO and TSS were considered as continuous variables. The stepwise feature selection process for the random forest (RF) [Citation31] was based on three steps [Citation31,Citation32]. First, elimination of variables with low importance by ranking the average of the variable importance measure on 50 runs of RF. Second, calculation of the Out-of-bag (OOB) error rates of 50 RF runs for each nested model (from the most important variable to the model with all variables previously selected). The variables included in the model with the lowest OOB error were selected. Third, selection of the final model by performing an ascending sequence of RF that tests the inclusion of each variable selected in the second step. The relationship between the selected miRNAs and the outcomes was evaluated using generalized additive models (GAMs) with penalized cubic regression splines. The same models were used to adjust miRNA levels for age, sex, previous pulmonary disease, smoking history and the use of corticoids after hospital discharge. A sparse inverse covariance matrix using a Lasso (L1) penalty was estimated to study the correlation between miRNAs selected [Citation33]. Correlations between continuous variables were estimated using Spearman rank correlation. Point-biserial correlations were used to calculate the correlations between dichotomous and continuous variables. The p-value threshold defining statistical differential expression was set at <0.05. All statistical analyses were performed using R software, version 4.0.2.
Results
Clinical characteristics of the patients
The study flowchart is presented in Supplemental Figure S1. Finally, 157 patients with ARDS secondary to SARS-CoV-2 infection and 33 patients who did not develop ARDS were available for miRNA quantification. Three samples were discarded due to insufficient plasma volume, the presence of hemolysis or low quality (Supplemental Figure S2A & S2B). Five miRNAs, miR-34b-5p, miR-34c-5p, miR-124-3p, miR-208a-3p and miR-208b-3p, were not considered in the subsequent analysis due to undetectable levels in more than 80% of the samples.
displays the characteristics of the study population. The median [P25;P75] age was 61 [53.2;66.8], and females represented 30.5% of the population. DLCO was abnormal (<80% predicted) in 81.8% of the population. The TSS score showed a median [P25;P75] of 5 [2;8]. The characteristics of the non-ARDS control groups are shown in Supplemental Table S2.
Table 1. Characteristics of the study population.
MicroRNA profiling of the pulmonary function
The characteristics of the study population according to DLCO tertiles are detailed in . Patients with low DLCO levels were older and had longer hospital and ICU stays, higher requirements for prone positioning and a higher lung affectation in the chest CT ( & Supplemental Figure S3).
Table 2. Characteristics of patients who entered in the inclusion criteria based on DLCO tertiles.
In univariate analyses, pulmonary function (as DLCO tertiles) was inversely associated with levels of miR-146a-5p [fold change (FC) = 0.849] (A). RF selected a miRNA profile associated with DLCO levels (as continuous variable) composed of five miRNAs: miR-17-5p, miR-27a-3p, miR-126-3p, miR-146a-5p and miR-495-3p (B). miRNAs established a dense correlation network (C). The individual relation between DLCO levels and the plasma miRNAs selected was analyzed using GAM modelling. miR-17-5p, miR-27a-3p and miR-146a-5p were inversely and lineally associated with DLCO (D); high levels of these three miRNAs correlated with low to very low levels of DLCO, i.e. diffusion impairment. Similar results were observed after adjustment for confounders: age, sex, previous pulmonary disease, smoking history and the use of corticoids after hospital discharge (Supplemental Figure S4A). These associations were not observed in non-ARDS patients positive for SARS-CoV-2 (Supplemental Figure S5A). No correlation was observed between miRNA levels and invasive mechanical ventilation (IMV) duration or hospital and ICU stays (Supplemental Figure S6).
Figure 1. Molecular mechanisms associated with pulmonary function in survivors of SARS-CoV-2-induced ARDS. A) Volcano plot representing the p-value versus the fold change for each microRNA after comparison of DLCO tertiles. The red dot indicates significant differences. B) Prediction model based on Random Forest. On the left, the importance of the contribution of each microRNA to the model. On the right, the best combination of microRNAs selected by the algorithm to reduce the error. C) Correlation between microRNAs that composed the signature. D) Linear or nonlinear relationship between the levels of each microRNA that composed the signature and DLCO. The expression levels are expressed as log10 (2−ΔCq) for statistical purposes. E) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (selected). F) Gene Ontology (GO) analysis (selected). The p-value denotes the significance of the molecular pathway or the biological process and the size of the points represents the number of genes involved. The false discovery rate adjusted p-value cutoff was 0.05.
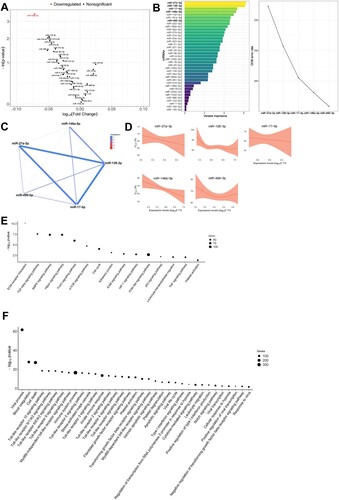
Pathway and GO term enrichment analyses were performed to further understand the biological implications of the miRNA profiles described. Sixty-one KEGG pathways and one hundred and fifty-one GO terms were enriched in experimentally validated target genes of the miRNAs that composed the signature (Supplemental Tables S3 and S4). The analyses identified molecular pathways and biological processes related to fibrosis (e.g. TGF-beta signalling pathway, mTOR signalling pathway, Fibroblast growth factor receptor signalling pathway), inflammation and immune response (e.g. Leukocyte transendothelial migration, TNF signalling pathway, several Toll-like receptor signalling pathways, Type I interferon signalling pathway, Cytokine-mediated signalling pathway, Leukocyte migration), cell death (e.g. Apoptotic signalling pathway), hypoxia (e.g. HIF-1 signalling pathway, Cellular response to hypoxia), vascularization (e.g. Notch signalling pathway) and coagulation (e.g. Platelet activation, Blood coagulation, Platelet degranulation) (E & F). Several molecular GO terms linked to viral infections were also identified (e.g. Viral process, Viral life cycle, Viral transcription, Positive regulation of viral transcription, Response to virus) (E & F).
MicroRNA profiling of the radiologic features
A similar workflow was used to analyze the relation between the radiologic features and the circulating miRNA profile. shows the characteristics of the study population according to TSS tertiles. Higher levels of TSS were directly associated with age, monocyte count, creatinine and urea levels at hospital admission, hospital stay, ICU stay, the use of IMV, IMV duration, use of prone positioning, and prone positioning duration ( & Supplemental Figure S7).
Table 3. Characteristics of patients who entered in the inclusion criteria based on TSS tertiles.
In univariate analyses, miR-9-5p [FC = 0.823], miR-16-5p [FC = 0.876] and miR-221-3p [FC = 0.864] were inversely associated with TSS (as tertiles) (A). Following the same approach, the feature selection procedure constructed a signature composed of four miRNAs: miR-9-5p, miR-21-5p, miR-24-3p and miR-221-3p (using TSS as continuous variable) (B). Except for miR-9-5p, which was poorly correlated with miR-221-3p, all miRNAs correlated with each other (C). GAM modelling showed that low levels of both miR-9-5p and miR-24-3p adjusted to an inverse linear relation with TSS and, consequently, higher levels of radiologic abnormalities. A nonlinear relationship was observed for miR-21-5p and miR-221-3p (D). Confounding factors showed no significant impact on the miRNA-TSS relationship (Supplemental Figure S4B). Except for miR-24-3p, the relationship between TSS levels and the selected miRNAs was only observed in the ARDS group (Supplemental Figure S5B). Again, no correlation was observed between IMV duration or hospital and ICU stays and miRNA levels (Supplemental Figure S6).
Figure 2. Molecular mechanisms associated with radiologic features in survivors of SARS-CoV-2-induced ARDS. A) Volcano plot representing the p-value versus the fold change for each microRNA after comparison of TSS tertiles. The red dots indicate significant differences. B) Prediction model based on Random Forest. On the left, the importance of the contribution of each microRNA to the model. On the right, the best combination of microRNAs selected by the algorithm to reduce the error. C) Correlation between microRNAs that composed the signature. D) Linear or nonlinear relationship between the levels of each microRNA that composed the signature and DLCO. The expression levels are expressed as log10 (2−ΔCq) for statistical purposes. E) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (selected). F) Gene Ontology (GO) analysis (selected). The p-value denotes the significance of the molecular pathway or the biological process and the size of the points represents the number of genes implicated. The false discovery rate adjusted p-value cutoff was 0.05.
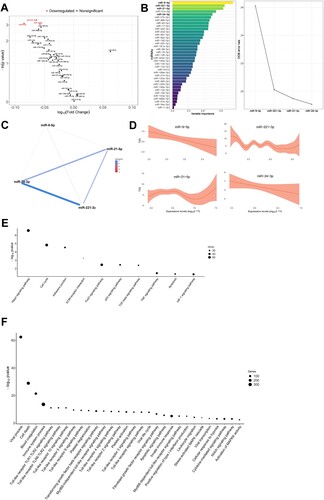
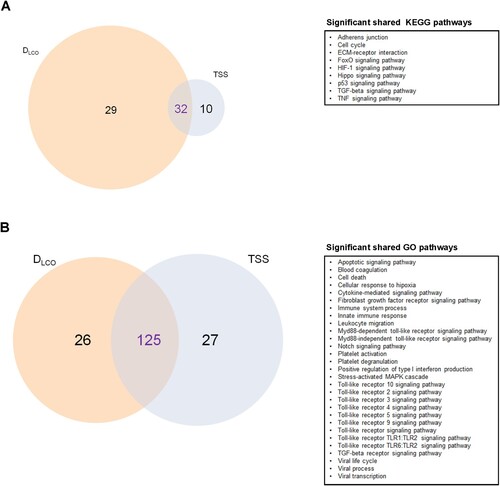
Forty-two KEGG pathways and 115 GO terms contained experimentally validated miRNA:gene interactions (Supplemental Tables S5 and S6). Again, the analyses identified molecular pathways and biological processes related to fibrosis (e.g. TGF-beta signalling pathway, Fibroblast growth factor receptor signalling pathway), inflammation and immune response (e.g. TNF signalling pathway, several Toll-like receptor signalling pathways, Leukocyte migration), cell death (e.g. Apoptotic signalling pathway), hypoxia (e.g. HIF-1 signalling pathway, Cellular response to hypoxia), vascularization (e.g. Notch signalling pathway) and coagulation (e.g. Blood coagulation, Platelet degranulation, Platelet activation) and viral infections (e.g. Viral process, Viral life cycle, Viral transcription) (E & F). An overlap between those molecular pathways and GO terms enriched in targets from the miRNA signatures associated with lung function and radiologic features was observed ().
Figure 3. Molecular mechanisms associated with pulmonary function and/or radiologic features. A) Venn diagram displaying the shared and unique Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (selected). B) Venn diagram displaying the shared and unique Gene Ontology (GO) terms (selected). The size of each circle is proportional to the total number of molecular pathways or biological processes related to DLCO and TSS. On the right, significant shared KEGG pathways and GO terms are reported for each diagram.
Molecular mechanisms involved in viral infection and response were highly enriched among the identified GO terms for both signatures. Therefore, we evaluated the presence of viral antigens in plasma samples from a subset of 50 survivors of SARS-CoV-2-induced ARDS. Antigenemia was presented in two patients (Supplemental Figure S8).
Discussion
Current study combined experimental and computational approaches to first evaluate the association between lung function and radiologic features and the circulating miRNA profile in survivors of SARS-CoV-2-induced ARDS 3 months after hospital discharge and then decipher the molecular mechanisms implicated in pulmonary outcomes. We report four major findings: i) distinct miRNA profiles are associated with pulmonary function and radiologic features; ii) this association seems to be specific for ARDS secondary to SARS-CoV-2 infection; iii) pulmonary function and structural features are linked not only to mechanisms such as fibrosis or inflammation but also to cell death, immune response, hypoxia, vascularization, coagulation and viral infection; iv) as such, the mechanisms linked to the respiratory sequelae of SARS-CoV-2-induced ARDS might implicate multiple biological pathways.
Individual miRNAs have a modest impact on the regulation of biological processes [Citation34]. Collective and coordinated miRNA expression networks, rather than single miRNAs, are critical for the regulation of the cellular phenotype [Citation13]. The analysis of miRNA profiles is paramount. Furthermore, the deregulation in individual expression programmes may not explain the complexity of the disease. For instance, Barbagallo et al. [Citation35] have recently demonstrated a profound perturbation in competitive endogenous RNA networks due to SARS-CoV-2 infection which ultimately impacts on disease progression. Advanced statistical tools seem fundamental to describe the interaction between miRNAs and the phenotype and identify nonlinear associations between miRNA levels and the outcome. Consequently, we constructed miRNA signatures using machine learning models. RF-based predictive models identified two miRNA signatures associated with pulmonary function and radiologic features in survivors of ARDS secondary to SARS-CoV-2 infection. The current results could improve the limited understanding of sequelae in survivors of SARS-CoV-2-induced ARDS. Supporting the significant functional redundancy among miRNAs [Citation36], although the individual miRNAs that composed the patterns were different, the downstream regulatory effects were highly similar with miR-17-5p, miR-27a-3p, miR-126-3p, miR-146a-5p and miR-495-3p for DLCO and miR-9-5p, miR-21-5p, miR-24-3p and miR-221-3p for TSS.
As expected, both pulmonary function and radiological features were closely linked to fibrotic and inflammatory pathways. The reduction in miR-9-5p, a suppressor of TGF-β1-dependent myofibroblast phenotypic transformation in lung disease [Citation37], may indicate a profibrotic state in survivors of severe forms of COVID-19. The results in subjects with persistent symptoms after acute COVID-19 [Citation38] and bioinformatic prediction using experimentally validated miRNA interactions corroborated this notion. Different cellular pathways related to fibrotic mechanisms in lung conditions were identified, including the AMPK signalling pathway [Citation39], Hippo signalling pathway [Citation40] and mTOR signalling pathway [Citation41]. Concerning inflammation, multiple KEGG pathways and GO terms, such as signalling pathways involving TNF or Toll-like receptors, were enriched in the target genes of the miRNA patterns. More specifically, the levels of miR-146a-5p were closely and inversely associated with lung function. This miRNA is induced in response to proinflammatory cytokines and participates in a negative feedback regulation loop to control inflammation [Citation42]. Therefore, its increase in survivors with higher pulmonary impairment may constitute an anti-inflammatory response to the extensive inflammatory insult inherent to the pathogenesis of ARDS, similar to the production of anti-inflammatory cytokines in proinflammatory conditions. In fact, hypercytokinemia is a characteristic feature of severe COVID-19 [Citation43], and its prolongation even 40–60 days postviral infection has been reported [Citation44]. The biological functions of the other miRNAs that composed the signatures support this hypothesis. In vitro and in vivo approaches suggest that mesenchymal stem cell-derived extracellular vesicles mitigate acute lung injury at least partially by transferring miR-27a-3p to alveolar macrophages, a regulator of NFKB1 and M2 macrophage polarization [Citation45]. Endothelial progenitor cell exosomes reduce local inflammatory cytokines in a model of lipopolysaccharide-induced acute lung injury, in part through the delivery of miR-126 [Citation46].
The circulating miRNA signature may be informative about other driving factors that mediate sequelae from SARS-CoV-2-induced ARDS. KEGG and GO analyses highlighted pathways related to platelet function and blood coagulation. SARS-CoV-2 infection induces a pro-coagulant state that has been recognized as a hallmark of COVID-19 [Citation47]. We have previously demonstrated alterations in the circulating profile of miRNAs implicated in coagulation mechanisms during the acute phase of the disease [Citation27]. According to our findings, the pathways related to the coagulation process remain perturbed even 3 months after hospital discharge. Although the clinical relevance remains uncertain [Citation48], these results are in line with the occurrence of thrombotic events after discharge or resolution of infection [Citation49]. In addition, miRNA signatures were enriched in targets of signalling pathways implicated in cell death/apoptosis (e.g. FoxO signalling pathway or p53 signalling pathway, among others), indicating that the cell injury mechanisms modulated during the acute phase by SARS-CoV-2 [Citation50] are still deregulated in “post-COVID” syndrome. Interestingly, the increased level of miR-126-3p in patients with significant diffusion impairment is compatible with aberrant angiogenic signalling. Cao et al. [Citation51] recently demonstrated that miR-126-3p inhibits the angiogenic function of human lung microvascular endothelial cells, which are essential for gas exchange and for lung injury repair and regeneration. Current results may have a relevant impact, since the mechanisms that mediate microvascular injury in ARDS sequelae are uncertain [Citation52]. Possible evidence for the phenotypic implications of our findings is given by the identification of target genes in hypoxia signalling pathways. The perturbations in these mechanisms may explain the decreased average, final and minimal oxygen saturation in the 6-minute walking test that we have previously described in survivors of SARS-CoV-2-induced ARDS [Citation6]. Of note, the miRNA signatures were also enriched in targets from GO terms related to viral infection. Despite this fact, we have not detected the virus in the blood through antigenemia profiling. A plausible explanation is the presence of viral fragments in other tissues causing immunoreactivity and persistent inflammation [Citation53]. The alteration of these biological processes during the recovery phase deserves additional investigation.
Although the endocrine function of circulating miRNAs is still not completely elucidated [Citation54], our work suggests a potential role of miRNAs in the respiratory sequelae of severe COVID-19. Overall, the current results provide a framework for targeted interventions against the sequelae of patients who develop SARS-CoV-2-induced ARDS. The management of the long-lasting effects of this syndrome should be based on a comprehensive and multidisciplinary approach to not only focus on inflammation and fibrosis but also other relevant pathological processes.
Our results should be considered in the context of several limitations. First, survivors of ARDS secondary to SARS-CoV-2 infection were recruited in a single centre. The results should be validated in independent cohorts. Second, potential confounding due to previous comorbidities, clinical management and inpatient complications should not be discarded. Third, how the deregulation of the miRNA signature in plasma impacts the pulmonary sequelae remains to be elucidated. Further functional studies may provide additional insights into role of the miRNAs in pulmonary abnormalities. Forth, we used a targeted approach using miRNAs previously associated with COVID-19 and/or pathological mechanisms linked to COVID-19/ARDS [Citation27]. Other miRNAs could be associated with pathophysiology of the postacute pulmonary sequelae. Large-scale miRNA analyses are fundamental. Fifth, potential overfitting should be acknowledged. In addition, the impact of type I error should not be discarded.
In conclusion, the pulmonary function and radiologic features in survivors of ARDS secondary to SARS-CoV-2 infection are associated with specific plasma miRNA patterns. The multifactorial mechanisms linked to the miRNA profiles provide novel knowledge of the physiopathology of persistent pulmonary dysfunction in the recovery stage and, in consequence, further insights into postacute care strategies.
Supplemental Material
Download MS Word (1.3 MB)Acknowledgments
The authors are indebted to María Arguimbau, Raquel Campo, Natalia Jarillo, Javier Muñoz and Manuel Sánchez(†) for their extensive support with project management and article preparation. This work was supported by IRBLleida Biobank (B.0000682) and “Plataforma Biobancos PT20/00021”. The human sample manipulation was performed in the Cell Culture Technical Scientific Service of the Universitat de Lleida (Lleida, Catalonia, Spain). The authors acknowledge all the COVID-19 patients and health workers involved in the study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Masclans J, Roca O, Muñoz X, et al. Quality of life, pulmonary function, and tomographic scan abnormalities after ARDS. Chest. 2011;139:1340–1346.
- Heyland D, Groll D, Caeser M. Survivors of acute respiratory distress syndrome: Relationship between pulmonary dysfunction and long-term health-related quality of life*. Crit Care Med. 2005;33:1549–1556.
- Ngai JC, Ko FW, Ng SS, et al. The long-term impact of severe acute respiratory syndrome on pulmonary function, exercise capacity and health status. Respirology. 2010;15:543–550.
- Ahmed H, Patel K, Greenwood DC, et al. Long-term clinical outcomes in survivors of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronavirus outbreaks after hospitalisation or ICU admission: A systematic review and meta-analysis. J Rehabil Med. 2020;52, jrm00063.
- Hui D, Joynt G, Wong KT, et al. Impact of severe acute respiratory syndrome (SARS) on pulmonary function, functional capacity and quality of life in a cohort of survivors. Thorax. 2005;60:401–409.
- González J, Benítez ID, Carmona P, et al. Pulmonary function and radiologic features in survivors of critical COVID-19. Chest. 2021;160:187–198.
- van Gassel RJJ, Bels JLM, Raafs A, et al. High prevalence of pulmonary sequelae at 3 months after hospital discharge in mechanically ventilated survivors of COVID-19. Am J Respir Crit Care Med. 2021;203:371–374.
- Blanco JR, Cobos-Ceballos MJ, Navarro F, et al. Pulmonary long-term consequences of COVID-19 infections after hospital discharge. Clin Microbiol Infect. 2021;27(6):892–896.
- Huang C, Huang L, Wang Y, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. The Lancet. 2021;397:220–232.
- Guler S, Ebner L, Aubry-Beigelman C, et al. Pulmonary function and radiological features 4 months after COVID-19: first results from the national prospective observational Swiss COVID-19 lung study. Eur Respir J. 2021;57(2003690.
- Sibila O, Albacar N, Perea L, et al. Lung function sequelae in COVID-19 patients 3 months after hospital discharge. Arch Bronconeumol. 2021;57:59–61.
- García-Hidalgo MC, González J, Benítez ID, et al. Proteomic profiling of lung diffusion impairment in the recovery stage of SARS-CoV-2–induced ARDS. Clin Transl Med. 2022;12(5):e838.
- Ebert MS, Sharp PA. Roles for MicroRNAs in conferring robustness to biological processes. Cell Elsevier B.V. 2012: 515–524.
- Mendell J, Olson E. Micrornas in stress signaling and human disease. Cell. 2012;148:1172–1187.
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659.
- Mitchell P, Parkin R, Kroh E, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518.
- Mori M, Ludwig R, Garcia-Martin R, et al. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab 2019;30:656–673.
- Cheng M, Yang J, Zhao X, et al. Circulating myocardial microRNAs from infarcted hearts are carried in exosomes and mobilise bone marrow progenitor cells. Nat Commun. 2019;10:959.
- Gomez I, Ward B, Souilhol C, et al. Neutrophil microvesicles drive atherosclerosis by delivering miR-155 to atheroprone endothelium. Nat Commun. 2020;11:214.
- Holtzman J, Lee H. Emerging role of extracellular vesicles in the respiratory system. Exp Mol Med. 2020;52:887–895.
- Torres A, Arguimbau M, Bermejo-Martín J, et al. Ciberesucicovid: un proyecto estratégico para una mejor comprensión y manejo clínico de la COVID-19 en pacientes críticos. Arch Bronconeumol. 2021;57:1–2.
- Ranieri VM, Rubenfeld GD, Thompson BT, et al. Acute respiratory distress syndrome: The Berlin definition. JAMA - J Am Med Assoc. 2012;307:2526–2533.
- Celli BR, MacNee W, Agusti A, et al. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J. 2004;23:932–946.
- Roca J, Burgos F, Sunyer J, et al. References values for forced spirometry. Eur Respir J. 1998;11:1354–1362.
- Ooi GC, Khong PL, Müller NL, et al. Severe acute respiratory syndrome: temporal lung changes at thin-section CT in 30 patients. Radiology. 2004;230:836–844.
- Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–815.
- de Gonzalo-Calvo D, Benítez ID, Pinilla L, et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl Res. 2021;S1931-5244:00122–00125.
- Blondal T, Jensby Nielsen S, Baker A, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods Academic Press Inc. 2013: S1–S6.
- Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Web Serv Issue Publ Online. 2015;43.
- Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47.
- Breiman L. Random forests. Mach Learn. 2001;45:5–32.
- Genuer R, Poggi JM, Tuleau-Malot C. Variable selection using random forests. Pattern Recognit Lett. 2010;31:2225–2236.
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432–441.
- Baek D, Villén J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71.
- Barbagallo D, Palermo CI, Barbagallo C, et al. Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells. Cell Mol Life Sci. 2022;79:75.
- Miska EA, Alvarez-Saavedra E, Abbott AL, et al. Most caenorhabditis elegans microRNAs Are individually Not essential for development or viability. PLOS Genet. 2007;3:e215.
- Fierro-Fernández M, Busnadiego Ó, Sandoval P, et al. Mir-9-5p suppresses pro-fibrogenic transformation of fibroblasts and prevents organ fibrosis by targeting NOX 4 and TGFBR 2. EMBO Rep 2015;16:1358–1377.
- Chun HJ, Coutavas E, Pine AB, et al. Immunofibrotic drivers of impaired lung function in postacute sequelae of SARS-CoV-2 infection. JCI Insight. 2021;6:e148476.
- Kheirollahi V, Wasnick RM, Biasin V, et al. Metformin induces lipogenic differentiation in myofibroblasts to reverse lung fibrosis. Nat Commun. 2019;10:1–16.
- Gokey JJ, Sridharan A, Xu Y, et al. Active epithelial hippo signaling in idiopathic pulmonary fibrosis. JCI Insight. 2018;3:e98738.
- Platé M, Guillotin D, Chambers RC. The promise of mTOR as a therapeutic target pathway in idiopathic pulmonary fibrosis. Eur Respir Rev. 2020;29:200269.
- Taganov KD, Boldin MP, Chang K-J, et al. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci. 2006;103:12481–12486.
- McElvaney OJ, McEvoy NL, McElvaney OF, et al. Characterization of the inflammatory response to severe COVID-19 illness. Am J Respir Crit Care Med. 2020;202:812–821.
- Heywood WE, Doykov I, Hällqvist J, et al. ‘The long tail of COVID-19’ - The detection of a prolonged inflammatory response after a SARS-CoV-2 infection in asymptomatic and mildly affected patients. F1000Res. 2020;9:1349.
- Wang J, Huang R, Xu Q, et al. Mesenchymal stem cell-derived extracellular vesicles alleviate acute lung injury Via transfer of miR-27a-3p∗. Crit Care Med. 2020;48:E599–E610.
- Zhou Y, Li P, Goodwin AJ, et al. Exosomes from endothelial progenitor cells improve outcomes of the lipopolysaccharide-induced acute lung injury. Crit Care. 2019;23:44.
- Malas MB, Naazie IN, Elsayed N, et al. Thromboembolism risk of COVID-19 is high and associated with a higher risk of mortality: A systematic review and meta-analysis. EClinicalMedicine. 2020;29-30:100639.
- Roberts LN, Whyte MB, Georgiou L, et al. Postdischarge venous thromboembolism following hospital admission with COVID-19. Blood. 2020;136:1347.
- Kanso M, Cardi T, Marzak H, et al. Delayed pulmonary embolism after COVID-19 pneumonia: a case report. Eur Hear J - Case Reports. 2020;4:1–4.
- Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761–770.
- Cao D, Mikosz AM, Ringsby AJ, et al. MicroRNA-126-3p inhibits angiogenic function of human lung microvascular endothelial cells via LAT1 (L-type amino acid transporter 1)-mediated mTOR (mammalian target of rapamycin) signaling. Arterioscler Thromb Vasc Biol. 2020;40:1195–1206.
- Katira BH, Giesinger RE, Engelberts D, et al. Adverse heart–lung interactions in ventilator-induced lung injury. Am J Respir Crit Care Med. 2017;196:1411–1421.
- Salamanna F, Veronesi F, Martini L, et al. Post-COVID-19 syndrome: The persistent symptoms at the post-viral stage of the disease. A systematic review of the current data. Front Med. 2021;0:392.
- Pinilla L, Benitez ID, González J, et al. Peripheral blood microRNAs and the COVID-19 patient: methodological considerations, technical challenges and practice points. RNA Biol.; 2021. p. 688–695.
