ABSTRACT
Carbapenem-resistant Pseudomonas aeruginosa (CRPA) has been a major threat to human health due to its increased morbidity and mortality in clinical settings. Carbapenemase genes are less frequently found in CRPA compared with carbapenem-resistant Enterobacterales, of which carbapenemase producers are common. In this study, we identified 11 blaKPC-2-harbouring P. aeruginosa isolates from 139 carbapenemase-insensitive P. aeruginosa isolates collected between 2010 and 2021 in a tertiary hospital in China. Nine isolates belonged to ST697, while the other two were ST463. The antibiotic susceptibility testing showed that all the isolates were multidrug resistant, including resistance to imipenem, meropenem, ceftazidime, and tigecycline. Patients with Klebsiella pneumoniae carbapenemase-2 (KPC-2)-producing P. aeruginosa infections were mostly associated with complicated diseases and prolonged hospital stay, with 30% deterioration. The whole-genome sequencing analysis showed that these isolates carried multiple antibiotic resistance genes and virulence genes, and the KPC-2 genetic elements were highly related in ST697 isolates. The complete sequencing of ST697 isolate SE5416 showed that the harbouring of blaKPC-2 resulted from complex transposition and homologous recombination of an IncpRBL16 plasmid and other mobile elements. The Galleria mellonella infection model experiment showed that these KPC-2-producing P. aeruginosa–infected larvae had low survival rates and high virulence. The present study revealed the shifting of CRPA from ST697 to ST463 in East China; ST463 had higher drug resistance, posing greater challenges for clinical management.
Introduction
Pseudomonas aeruginosa is one of the most common Gram-negative bacteria in China [Citation1]. P. aeruginosa can easily colonize in the human body and invade the respiratory tract, urinary tract, surgical site, blood, abdominal cavity, and skin/soft tissues, causing hospital-acquired pneumonia [Citation2,Citation3]. P. aeruginosa has the characteristics of high mutation rates with complex antibiotic resistance mechanisms [Citation4]. As an opportunistic pathogen, it is one of the main causes of morbidity and death in patients with cystic fibrosis and individuals with weakened immune functions [Citation5]. In the last two decades, the spread of multidrug-resistant (MDR) P. aeruginosa, including those resistant to third-generation cephalosporins and carbapenem antibiotics, has greatly increased worldwide [Citation6,Citation7].
Carbapenems are important antibiotics for treating MDR P. aeruginosa infections [Citation8]. Unlike carbapenem-resistant Enterobacterales in which the carbapenem resistance is mainly caused by the production of different carbapenemases (Ambler class A, B, and D), carbapenem resistance in P. aeruginosa is more commonly associated with intrinsic resistance mechanisms, including overexpression of the efflux pump system, overexpression of chromosomal cephalosporinase, and reduction or loss of OprD outer membrane protein expression [Citation9]. In addition, among clinical carbapenemase-producing P. aeruginosa (CPPA), Ambler class B metal-β-lactamases (MBLs, e.g. VIM and IMP) were more commonly reported, whereas class A carbapenemases [e.g. Klebsiella pneumoniae carbapenemase (KPC)] were less frequently described, especially in China [Citation10]. In 2007, scientists from Columbia first discovered a P. aeruginosa isolate harbouring the blaKPC-2 gene [Citation11]; since then, additional reports on such isolates in other foreign countries have been published [Citation12,Citation13]. Subsequent studies found that the blaKPC gene was mobilized on the 10-kb Tn3-family active transposon Tn4401, which was delimited by two 39-bp inverted-repeat sequences [Citation14]. Currently, ST235, ST111, and ST175 are high-risk P. aeruginosa clones in clinical isolates worldwide, often associated with multidrug resistance or extensively drug resistance [Citation6].
The present 12-year retrospective study was conducted in a tertiary hospital, and 11 KPC-2-producing P. aeruginosa isolates were identified. Most of these isolates belonged to an uncommon ST697 clone, whereas the remaining recently isolated clones belonged to the emerging high-risk clone ST463. In vitro phenotypic experiments and in vivo Galleria mellonella infection model results suggested that these KPC-2-producing P. aeruginosa isolates were highly resistant and virulent, and required more attention and further research for their prevention and control.
Materials and methods
Collection of isolates
A total of 139 P. aeruginosa isolates that were not sensitive to carbapenem antibiotics (minimum inhibitory concentration of meropenem ≥4 μg/mL and/or imipenem ≥4 μg/mL) were collected from the Second Affiliated Hospital of Soochow University from 2010 to 2021. All isolates were speciated using the MALDI-TOF MS apparatus (Bruker Microflex LT, Bruker Daltonik GmbH, Bremen, Germany). The isolates were cultured using lysogeny broth (LB) solid medium (pH = 7.0) with subinhibitory meropenem/imipenem concentration (0.5 μg/mL) at 37°C.
Molecular detection
BlaKPC was screened using polymerase chain reaction, followed by Sanger sequencing [Citation15]. The DNA template was prepared by the boiling method. In brief, a loopful of bacteria were added to 100 μL of ddH2O and boiled for 10 min, followed by centrifugation at 5000 rpm and 4°C for 5 min. The supernatant was then diluted at a ratio of 1:10 in fresh ddH2O and used as a DNA template.
Antibiotic sensitivity testing
The antimicrobial susceptibility testing was performed using the standard broth microdilution method, and the results were interpreted following the 2020 CLSI breakpoints [Citation16]. The experiment was performed in three biological replicates on two different dates. P. aeruginosa ATCC27583 was used as the quality control.
Genome sequencing analysis
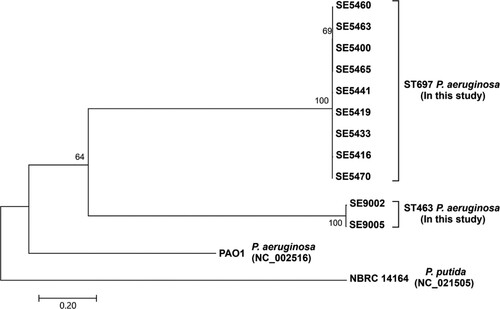
Whole-genome sequencing was performed on all KPC-2-producing P. aeruginosa isolates. The maximum-likelihood phylogenetic tree of KPC-producing P. aeruginosa isolates based on recombination-free core genome single-nucleotide polymorphisms (SNPs) was conducted with MUMmer 3.0, ClonalFrameML, and MEGA7, as described in a previous study [Citation17]. The sequence of P. aeruginosa PAO1 (GenBank accession no. NC_002516) was used as the reference, and the sequence of Pseudomonas putida NBRC 14164 (GenBank accession no. NC_021505) was used as the out-group.
G. mellonella infection model
All isolates were grown in 1 mL of LB liquid medium (pH = 7.0) and cultured with shaking at 200 rpm overnight at 37°C. The next day, the cultures were diluted at a ratio of 1:200 in 10 mL of fresh LB liquid medium (pH = 7.0) and grown for another 1.5–2 h at 37°C to mid-log phase (∼108 CFU/mL, 0.3 OD at 600 nm). Then, all cultures were washed and adjusted to ∼105 CFU/mL with normal saline before infection. The culture was then injected through the last left pro-leg with 10 μL of bacteria (103 CFU) [Citation18]. Preliminary experiments showed that the concentration of bacteria could better reflect the difference in virulence in these isolates. After infection, the number of survivors was recorded at 37°C for three consecutive days. A saline control group was included, and 20 larvae were used in the control and experimental groups. This experiment was divided into three groups: the experimental group, the normal saline group, and the blank control group. This experiment was performed in three biological replicates on two different dates.
Ethical approval
The ethical approval was obtained from the ethics committee of the Second Affiliated Hospital of Soochow University. The isolates used in this study were collected previously from routine microbiological specimens, while all the microbiological specimens were anonymized. The patients were not physically involved in this study. Therefore, no consent was needed for this study.
Results
Clinical and molecular characteristics of the isolates
A total of 139 clinical carbapenemase-insensitive P. aeruginosa isolates were collected from the Second Affiliated Hospital of Soochow University between 2010 and 2021. Molecular testing showed that 11 isolates harboured the blaKPC-2 gene. These isolates were recovered from the respiratory tract (n = 7, 63.6%), skin/soft tissue (n = 2, 18.2%), and urinary tract (n = 2, 18.2%). Among the patients, 70.0% (n = 7) were male and 30.0% (n = 3) were female (two isolates were recovered from the same patient during different periods). The median age was 58 years. Seven (70.0%) patients improved upon their discharge, while the condition of three (30%) of them deteriorated ().
Table 1. Information and molecular characteristics of the isolates.
Information on the medical history of seven patients showed that all patients had a long hospital stay of more than 30 days, with the longest being 110 days; five patients had received surgical treatment. These patients suffered from a variety of diseases, were in poor health conditions, and had been hospitalized in the intensive care unit (Table S1). They all received invasive ventilation and central venous catheterization adjuvant therapy, and three of them also received closed thoracic drainage (data not shown).
Antimicrobial susceptibility
The antibiotic susceptibility testing of these 11 KPC-2-P. aeruginosa isolates showed that they were nonsensitive to carbapenem antibiotics (meropenem and/or imipenem) and highly resistant to most antibiotics tested (). Ten of these isolates showed resistance rates of more than 85% to the 23 antibiotics tested, with most of them displaying resistance rates of more than 90%. The results showed that the resistance rates of these isolates to cefazolin, cefuroxime ceftriaxone, nitrofurantoin, tetracycline, minocycline, tigecycline, chloroamphenicol, ampicillin–sulbactam, and amoxicillin-clavulanic acid were 100%, while the resistance rates to tobramycin and amikacin were the lowest, 27.3% and 18.2%, respectively. However, two recently obtained isolates (SE9002 and SE9005) showed resistance to both tobramycin and amikacin, suggesting the emergence of the extensively resistant ST463 KPC-2-P. aeruginosa.
Table 2. In vitro susceptibility of different clinical antibiotics against 11 KPC-2-P. aeruginosa isolates.
Genome sequencing analysis
The genome sequencing results showed that the 11 KPC-2-P. aeruginosa clinical isolates harboured multiple antibiotic resistance genes (). Among these, qnrS2 and crpP conferred quinolone antibiotic resistance [Citation19]. CmlA1, catB7, fosA, aac(3)-IId, and aph(3’)-IIb encoded resistance to chloramphenicol, fosfomycin, and aminoglycosides [Citation20,Citation21]. Tet(G) was related to tetracycline resistance [Citation22,Citation23]. Sul1 was related to sulfa antibiotic resistance [Citation24]. In addition, genes such as sul2, armA, and mphA were located in the bacterial resistance determinant cluster (AbGRI1 and AbGRI3) [Citation25,Citation26].
Table 3. Whole genome sequencing results of antibiotic resistance genes and plasmids.
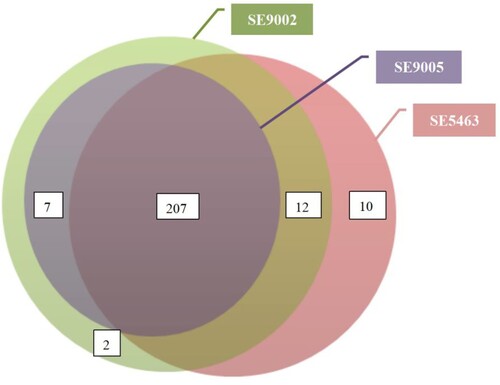
In silico screening showed that the isolates harboured more than 200 virulence genes. The total number of each was as follows: SE5463, SE5465, SE5470, SE5460, SE5441, SE5433, SE5416, SE5419, and SE5400 (n = 229); SE9002 (n = 228); and SE9005 (n = 214) (). These virulence genes primarily encode function or pathway of type II, III and VI secretion systems, flagellar and pili biosynthesis, exopolysaccharide, siderophore and lipopolysaccharide formation and secration etc., contributing to the invasion and destruction of host tissues (Table S2). All these genes were chromosome borne but were not carried by antibiotic resistance plasmids.
Figure 1. The relationship between the number of virulence genes carried by the isolates. The virulence genes of the remaining 8 isolates were consistent with SE5463, so they were not shown here.
The phylogenetic analysis of blaKPC-2 isolates showed that the ST697 and ST463 isolates formed two separate clusters (). The 9 ST697 isolates differed with an average of 26 core SNPs (range 5–45), while 2 ST463 isolates differed with an average of 78 core SNPs, suggesting that the spread of CPPA in the hospital was mainly associated with the clonal expansion of 2 KPC-2-producing P. aeruginosa isolates.
IncpRBL16 plasmid pSE5416-KPC
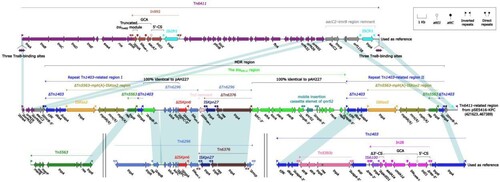
SE5416 was selected for complete genome sequencing using Illumina and PacBio RSII. The blaKPC-2 was harboured by a 510.71-kb IncpRBL16 plasmid, which was assigned the name pSE5416-KPC (the eight isolates of ST697 carry the similar plasmid) (data not shown). A detailed genetic dissection analysis was applied to this plasmid. The modular structure of pSE5416-KPC was divided into the backbone and eight accessory modules (defined as exogenous DNA regions inserted at different sites of the backbone) (). The backbone of pSE5416-KPC shared 98.56% nucleotide identity and 96% coverage with the IncpRBL16 reference plasmid pRBL16 (GenBank accession number CP015879) [Citation27]. Among the eight accessory modules pSE5416-KPC carried, only the Tn6411-related region harboured antimicrobial resistance genes [Citation28], mediating resistance to aminoglycosides (aacC2), β-lactams (blaKPC-2), fluoroquinolones (qnrS2), and macrolides (mph(A)). The Tn6411-related region could be preliminarily divided into several truncated Tn6411 fragments and a 41.99-kb MDR region, which was embedded in one of the truncated Tn6411 fragments (neatly carrying the aacC2 gene) with 5-bp direct repeats (DRs; target site duplication signals for transposition) (). The MDR region was composed of two repeat Tn1403-related regions [carrying the mph(A) gene] [Citation29], and the Tn6296-related blaKPC-2 region (carrying the blaKPC-2 and qnrS2 genes) [Citation30] inserted between two repeat regions with 8-bp DRs. Additionally, the blaKPC-2 region contained a partial plasmid sequence 100% identical to the IncU plasmid pAH227 with 36% coverage (GenBank accession number KT315926) [Citation31]. This was a new blaKPC-2 surrounded by △IS Kpn6 and IS Kpn27. This structure indicated that the harbouring of blaKPC-2 in pSE5416-KPC resulted from the complex transposition and homologous recombination of an IncpRBL16 plasmid and other mobile elements. Additionally, blaKPC-2 genes from SE9002 and SE9005 were located on chromosome. Further analysis showed that both these two blaKPC-2 genes were also carried by the Tn6296-related blaKPC-2 regions, which were highly similar to the one from pSE5416-KPC (data not shown).
Figure 3. Schematic diagram of the plasmid pSE5416-KPC. Genes of different functions are denoted by arrows and presented in various colours. The circles show (from outside to inside): predicted coding sequences, scale, backbone (black) and accessory module (gray) regions, GC content and GC skew [(G−C)/(G + C)].
![Figure 3. Schematic diagram of the plasmid pSE5416-KPC. Genes of different functions are denoted by arrows and presented in various colours. The circles show (from outside to inside): predicted coding sequences, scale, backbone (black) and accessory module (gray) regions, GC content and GC skew [(G−C)/(G + C)].](/cms/asset/704a3c95-e941-407f-8a5d-3bd7543d8d36/temi_a_2140609_f0003_oc.jpg)
Figure 4. Linear Comparison of Tn6411-related region and related regions. Genes are denoted by arrows. Genes, mobile elements, and other features are coloured based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within the plasmid pSE5416-KPC. The accession number of Tn64112, Tn14033, Tn62964, and Tn55636 used as reference are CP024477, AF313472, FJ628167, and U88088, respectively.
Survival rates of G. mellonella infection
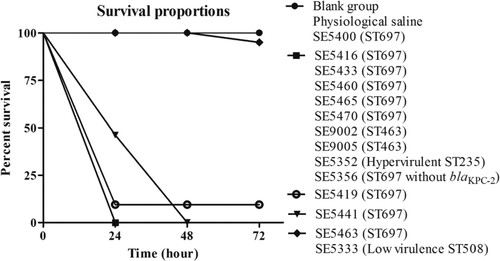
We selected G. mellonella larvae of similar weight (190–220 mg) to minimize the differences in body weight and standardize the age of the larvae to evaluate the virulence level of these isolates [Citation32]. We injected 103 CFU of different-numbered clinical isolates into the larvae and monitored the survival of the larvae every 24 h [Citation33].
Isolate SE5352 was an ST235 P. aeruginosa carrying exoU virulence gene, and SE5356 was an ST697 P. aeruginosa without blaKPC-2 (carrying 227 virulence genes, data not shown), collected by our research group during the same period. The exoU-harbouring ST235 P. aeruginosa was often accompanied by high virulence [Citation34]. The SE5333 selected in the experiment was an ST508-type P. aeruginosa without major virulence genes. SE5352 and SE5333 were used as the high- and low-virulence controls in the G. mellonella infection model.
The results showed that, compared with the two control groups (blank group and physiological saline group) and the low-virulence group (ST508–SE5333). The larvae infected by eight isolates, including ST697 isolates (SE5416, SE5433, SE5460, SE5465, SE5470, and SE5356), ST463 isolates (SE9002 and SE9005), and hypervirulent ST235 isolates (SE5352) had a survival rate of 0% after 24 h. The survival rate of larvae infected by ST697 SE5441 was 0% after 48 h, and that by ST697 SE5419 was 10% after 72 h. On the contrary, the survival rates of larvae infected by ST697 SE5400 and SE5463 were as high as 100% and 95% after 72 h, respectively. The low-virulence control strain ST508 SE5333 had the same survival rate as ST697 SE5463 (). The results suggested a higher virulence in the ST697 and ST463 P. aeruginosa isolates, and the high virulence of these clinically isolated KPC-2-P. aeruginosa had strong harmfulness.
Discussion
Today, P. aeruginosa nosocomial infections have become a global healthcare problem because they are largely associated with hospital-acquired infections, including ventilator-associated pneumonia, central catheter–associated bloodstream infections, catheter-associated infections, and surgical/transplant infections [Citation35,Citation36]. Especially in adults with cystic fibrosis, P. aeruginosa colonization ranged from 31% to 48%, and the development of antimicrobial resistance made it increasingly difficult to treat and eradicate [Citation37]. The overall mortality rate of P. aeruginosa bacteremia was more than 30% [Citation38,Citation39], the resistance rate to one or more antibiotic groups was 38% [Citation40], and the sensitivity to carbapenems decreased [Citation41]. Importantly, carbapenem-resistant bacterial infections caused by Acinetobacter baumannii or P. aeruginosa accounted for 82.3% of all Gram-negative bacteria [Citation42], which should not be underestimated. In addition, P. aeruginosa showed a higher propensity of mutations compared with other Gram-negative bacterial species, making P. aeruginosa more difficult to treat. The World Health Organization has classified Carbapenem-resistant Pseudomonas aeruginosa (CRPA) as one of the pathogens for which new alternative antibiotics are urgently required [Citation43]. At present, KPC-producing P. aeruginosa isolates are alarmingly increasingly worldwide, which has aroused the attention and vigilance of clinical researchers [Citation44–46].
In this study, 11 blaKPC-2-harbouring P. aeruginosa isolates and other resistance genes were identified from the 139 carbapenemase-insensitive P. aeruginosa isolates collected in China from 2010 to 2021. These KPC-2-P. aeruginosa isolates were collected from different clinical sites of the respiratory tract, skin/soft tissue, and urinary tract, with a large time span (). Different from the dominant clones ST235 and ST111 found in previous global studies [Citation47], we found the evolutionary trend of KPC-producing hypervirulent P. aeruginosa superior clone ST697 (first report) to ST463, which has become a potential high-risk clone in China in recent years [Citation48]. Not to be underestimated, these isolates were all MDR-PA, resulting in refractory patients with a mortality rate of up to 30% (). KPC β-lactamases, encoded by blaKPC-2, hydrolyze β-lactams of all classes and are highly efficient in hydrolyzing carbapenem antibiotics [Citation49]. The shifting of blaKPC-2-positive carbapenemase-insensitive P. aeruginosa isolates in this study significantly increased the level of resistance to carbapenems, other β-lactams and quinolones, especially the recently isolated ST463 strains that showed higher antibiotic resistance rates, resulting in greater challenges to treatment; hence, it should be closely monitored in hospitals in the future (, Table S3).
Besides intrinsic resistance and chromosomal mutations, mobile genetic elements such as plasmids and integrating conjugative elements are responsible for transmitting resistance genes to P. aeruginosa strains [Citation50]. Further whole-genome sequencing and analyis found that all these 11 isolates harboured multiple antibiotic resistance genes, leading to the ineffective treatment using antibiotics such as quinolone, aminoglycoside, tetracycline, and sulfa (), as demonstrated by the clinical antibiotic susceptibility testing results. The structure analysis indicated that the harbouring of blaKPC-2 in pSE5416-KPC resulted from the complex transposition and homologous recombination of an IncpRBL16 plasmid and other mobile elements ( and ). The antibiotic resistance gene blaKPC is most commonly harboured by plasmids. The NCBI GenBank database has listed 17 complete P. aeruginosa plasmids encoding KPC, of which three have been reported in China [Citation51]. IncpRBL16 is a 370.3-kb plasmid first reported in Pseudomonas citellella SJTE-3, which was isolated from the activated sludge from a sewage treatment plant in China, but did not carry the resistance gene blaKPC-2 [Citation52]. One of the most important vectors for the widespread blaKPC-2 gene transfer is Tn6296, which was originally discovered in the MDR plasmid pKP048 from Klebsiella pneumoniae [Citation51]. In this study, complex transposition and homologous recombination of Tn6296 and other related mobile elements promoted the expression of blaKPC-2 in the isolates, and was involved in influencing antibiotic sensitivity and even clinical symptoms of patients.
A variety of virulence factors lead to the pathogenesis of P. aeruginosa infection, including a variety of toxins, motor systems, and pigments. One of the most relevant P. aeruginosa virulence factors is the type III secretion system, which can directly transfer effector toxins (ExoT, ExoY, ExoS, and ExoU) into host cells [Citation53]. The clinically isolated hypervirulent KPC-2-P. aeruginosa in this study carried exoT, exoY, and exoS genes, and the infected G. mellonella larvae had low survival rates. Surprisingly, the 24-h mortality rates of infected larvae of ST463 isolates were 100% compared with those of ST697 isolates (56%) (), leading to severe and complex clinical symptoms and even poor clinical outcomes. Among these, exoU has been associated with a high-virulence phenotype and poor prognosis in patients with pneumonia and bacteremia [Citation54]. This may be because ExoU-positive P. aeruginosa isolates are more likely to resist multiple antibiotics, such as carbapenems, cephalosporins, fluoroquinolones, and aminoglycosides, which can exacerbate infection and increase mortality. Interestingly, a previous study reported exoU and exoS as mutually exclusive in P. aeruginosa [Citation48]. ST235, a high-risk MDR clone worldwide, highly correlated with exoU [Citation55], which was also the largest proportion (9%) of multilocus sequence typing in this study (Figure S1), all carrying exoU gene (data not shown). The clone of ST235/ST463 (with blaKPC-2) carrying exoU/exoS gene appeared and spread in East China instead of ST697, with higher virulence, and may become a new clinical threat. Althoug most of these virulence genes were found to be on the chromosome, the possibility of plasmid-borne virulence factors can not be ruled out. For example, the 510.71 kb pSE5416-KPC plasmid harbours over 637 genes, and accounts for ∼5% total P. aeruginosa genomes, and could potentially contain uncharacterized virulence factors that contribute to the increased virulence. The plasmid-borne virulence factors in P. aeruginosa deserves further studies.
Currently, treatment options for KPC-2-producing CRPA infections are very limited. Some novel antibiotics, such as ceftazidime-avibactam, ceftolozane-tazobactam, cefiderocol, and imipenem-cilastatin/relebactam showed promising activity against CRPA in certain studies [Citation56]. However, resistance to these agents have already emerged. For example, a recent study described the emergence of ceftazidime-avibactam resistant CRPA ST463 isolates in China, likely as a consequence of treatment selection pressure [Citation51]. Adequate empiric and definitive therapeutic decisions should be carefully considered during CRPA treatment to minimize selected resistance. Additional antibiotic development is urgently needed to provide sufficient options to successfully manage these CRPA infections.
This study was novel in reporting KPC-2-producing ST697-type P. aeruginosa, which was associated with high virulence in the G. mellonella infection model. KPC-2-producing P. aeruginosa ST463 was first reported in China in 2015 and has rapidly spread in Zhejiang province in recent years [Citation57]. In this study, we analyzed KPC-2-producing carbapenemase-insensitive P. aeruginosa isolates from 2010 to 2021 in East China and revealed that the shifting of CRPA from ST697 to ST463, which was more antibiotic resistant, posing greater challenges for clinical management, resulting in poor therapeutic efficacy. The clinical management should be alert to the emergence and spread of these MDR and potentially high virulent P. aeruginosa strains.
Supplemental Material
Download MS Word (131.7 KB)Acknowledgments
We are grateful for professor Dongsheng Zhou from State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology for the assistance in the drawing of figures.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hu FP, Guo Y, Zhu DM, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect. 2016;22 Suppl 1:S9–14.
- Micek ST, Wunderink RG, Kollef MH, et al. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: impact of multidrug resistance. Crit Care. 2015;19:219.
- Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39.
- Pacheco T, Bustos-Cruz RH, Abril D, et al. Pseudomonas aeruginosa coharboring BlaKPC-2 and BlaVIM-2 carbapenemase genes. Antibiotics (Basel). 2019;8(3):98.
- Lee AC, Jones AL. Multi-resistant Pseudomonas aeruginosa ST235 in cystic fibrosis. Paediatr Respir Rev. 2018;27:18–20.
- Oliver A, Mulet X, Lopez-Causape C, et al. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;21-22:41–59.
- McCarthy K. Pseudomonas aeruginosa: evolution of antimicrobial resistance and implications for therapy. Semin Respir Crit Care Med. 2015;36(1):044–055.
- Riera E, Cabot G, Mulet X, et al. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J Antimicrob Chemother. 2011;66(9):2022–2027.
- Castanheira M, Deshpande LM, Costello A, et al. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother. 2014;69(7):1804–1814.
- Rossi Goncalves I, Dantas RCC, Ferreira ML, et al. Carbapenem-resistant Pseudomonas aeruginosa : association with virulence genes and biofilm formation. Braz J Microbiol. 2017;48(2):211–217.
- Villegas MV, Lolans K, Correa A, et al. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother. 2007;51(4):1553–1555.
- Akpaka PE, Swanston WH, Ihemere HN, et al. Emergence of KPC-producing Pseudomonas aeruginosa in Trinidad and Tobago. J Clin Microbiol. 2009;47(8):2670–2671.
- Robledo IE, Aquino EE, Vazquez GJ. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob Agents Chemother. 2011;55(6):2968–2970.
- Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother. 2011;55(11):5370–5373.
- Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123.
- CLSI. Performance standards for antimicrobial susceptibility testing, 30th edition (M100S). Wayne (PA): CLSI; 2020.
- Liang Q, Jiang X, Hu L, et al. Sequencing and genomic diversity analysis of IncHI5 plasmids. Front Microbiol. 2019;9:3318.
- Cotter G, Doyle S, Kavanagh K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol. 2000;27(2):163–169.
- Chavez-Jacobo VM, Hernandez-Ramirez KC, Romo-Rodriguez P, et al. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob Agents Chemother. 2018;62(6):e02629-17.
- Ito R, Mustapha MM, Tomich AD, et al. Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. mBio. 2017;8(4):e00749-17.
- Zhu X, Li P, Qian C, et al. Prevalence of aminoglycoside resistance genes and molecular characterization of a novel gene, aac(3)-IIg, among clinical isolates of the Enterobacter cloacae complex from a Chinese teaching hospital. Antimicrob Agents Chemother. 2020;64(9):e00852-20.
- Sharma VK, Johnson N, Cizmas L, et al. A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere. 2016;150:702–714.
- Gao P, Xu W, Ruan X, et al. Long-term impact of a tetracycline concentration gradient on the bacterial resistance in anaerobic-aerobic sequential bioreactors. Chemosphere. 2018;205:308–316.
- Ozkaya E, Aydin F, Bayramoglu G, et al. Investigation of integrons, sul1-2 and dfr genes in trimethoprim-sulfametoxazole-resistant Stenotrophomonas maltophilia strains isolated from clinical samples. Mikrobiyol Bul. 2014;48(2):201–212.
- Blackwell GA, Holt KE, Bentley SD, et al. Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J Antimicrob Chemother. 2017;72(4):1031–1039.
- Rafaque Z, Dasti JI, Andrews SC. Draft genome sequence of a uropathogenic Escherichia coli isolate (ST38 O1:H15) from Pakistan, an emerging multidrug-resistant sequence type with a high virulence profile. New Microbes New Infect. 2019;27:1–2.
- Jiang X, Yin Z, Yuan M, et al. Plasmids of novel incompatibility group IncpRBL16 from Pseudomonas species. J Antimicrob Chemother. 2020;75(8):2093–2100.
- Zhan Z, Hu L, Jiang X, et al. Plasmid and chromosomal integration of four novel blaIMP-carrying transposons from Pseudomonas aeruginosa, Klebsiella pneumoniae and an Enterobacter sp. J Antimicrob Chemother. 2018;73(11):3005–3015.
- Stokes HW, Elbourne LD, Hall RM. Tn1403, a multiple-antibiotic resistance transposon made up of three distinct transposons. Antimicrob Agents Chemother. 2007;51(5):1827–1829.
- Wang D, Zhu J, Zhou K, et al. Genetic characterization of novel class 1 Integrons In0, In1069 and In1287 to In1290, and the inference of In1069-associated integron evolution in Enterobacteriaceae. Antimicrob Resist Infect Control. 2017;6:84.
- Dobiasova H, Videnska P, Dolejska M. Complete sequences of IncU plasmids harboring quinolone resistance genes qnrS2 and aac(6’)-Ib-cr in Aeromonas spp. from ornamental fish. Antimicrob Agents Chemother. 2016;60(1):653–657.
- Ramarao N, Nielsen-Leroux C, Lereclus D. The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J Vis Exp. 2012;70:e4392.
- Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7(3):214–229.
- Sanchez-Diener I, Zamorano L, Lopez-Causape C, et al. Interplay among resistance profiles, high-risk clones, and virulence in the caenorhabditis elegans Pseudomonas aeruginosa infection model. Antimicrob Agents Chemother. 2017;61(12):e01586-17.
- Nathwani D, Raman G, Sulham K, et al. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3(1):32.
- Rosenthal VD, Al-Abdely HM, El-Kholy AA, et al. International Nosocomial Infection Control Consortium report, data summary of 50 countries for 2010-2015: device-associated module. Am J Infect Control. 2016;44(12):1495–1504.
- Reece E, Segurado R, Jackson A, et al. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: an Irish registry analysis. BMC Pulm Med. 2017;17(1):70.
- Callejas-Diaz A, Fernandez-Perez C, Ramos-Martinez A, et al. Impacto de la bacteriemia por Pseudomonas aeruginosa en un hospital de tercer nivel: mortalidad y factores pronósticos. Med Clin (Barc). 2019;152(3):83–89.
- McCarthy KL, Paterson DL. Long-term mortality following Pseudomonas aeruginosa bloodstream infection. J Hosp Infect. 2017;95(3):292–299.
- Rosanova MT, Mussini MS, Arias AP, et al. Epidemiological features and risk factors for mortality in Pseudomonas aeruginosa bacteremia in children. Arch Argent Pediatr. 2019;117(2):128–131.
- Gaete ME, Valenzuela MP, Bachero AW, et al. Carbapenemasas en Pseudomonas aeruginosa con susceptibilidad disminuida a los carbapenémicos después de una década, desde VIM a KPC. Rev Chilena Infectol. 2020;37(4):389–394.
- Cai B, Echols R, Magee G, et al. Prevalence of carbapenem-resistant gram-negative infections in the United States predominated by acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infect Dis. 2017;4(3):ofx176.
- Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327.
- Wang LJ, Chen EZ, Yang L, et al. Emergence of clinical Pseudomonas aeruginosa isolate Guangzhou-PaeC79 carrying crpP, blaGES-5, and blaKPC-2 in Guangzhou of China. Microb Drug Resist. 2021;27(7):965–970.
- Yuan M, Guan H, Sha D, et al. Characterization of blaKPC-2-carrying plasmid pR31-KPC from a Pseudomonas aeruginosa strain isolated in China. Antibiotics (Basel). 2021;10(10):1234.
- Cai H, Zhu Y, Hu D, et al. Co-harboring of novel blaKPC–2 plasmid and integrative and conjugative element carrying Tn6203 in multidrug-resistant Pseudomonas aeruginosa. Front Microbiol. 2021;12:674974.
- Feng W, Huang Q, Wang Y, et al. Changes in the resistance and epidemiological characteristics of Pseudomonas aeruginosa during a ten-year period. J Microbiol Immunol Infect. 2021;54(2):261–266.
- Hu Y, Peng W, Wu Y, et al. A potential high-risk clone of Pseudomonas aeruginosa ST463. Front Microbiol. 2021;12:670202.
- Hu Y, Liu C, Wang Q, et al. Emergence and expansion of a carbapenem-resistant Pseudomonas aeruginosa clone are associated with plasmid-borne bla KPC-2 and virulence-related genes. mSystems. 2021;6(3):e00154-21.
- Botelho J, Grosso F, Peixe L. Antibiotic resistance in Pseudomonas aeruginosa - mechanisms, epidemiology and evolution. Drug Resist Updat. 2019;44:100640.
- Zhu Y, Chen J, Shen H, et al. Emergence of ceftazidime- and avibactam-resistant Klebsiella pneumoniae carbapenemase-producing Pseudomonas aeruginosa in China. mSystems. 2021;6(6):e0078721.
- Zheng D, Wang X, Wang P, et al. Genome sequence of pseudomonas citronellolis SJTE-3, an estrogen- and polycyclic aromatic hydrocarbon-degrading bacterium. Genome Announc. 2016;4(6):e01373-16.
- Sanchez-Diener I, Zamorano L, Pena C, et al. Weighting the impact of virulence on the outcome of Pseudomonas aeruginosa bloodstream infections. Clin Microbiol Infect. 2020;26(3):351–357.
- Pena C, Cabot G, Gomez-Zorrilla S, et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis. 2015;60(4):539–548.
- Cholley P, Ka R, Guyeux C, et al. Population structure of clinical Pseudomonas aeruginosa from West and Central African countries. PLoS One. 2014;9(9):e107008.
- Kazmierczak KM, de Jonge BLM, Stone GG, et al. In vitro activity of ceftazidime/avibactam against isolates of Pseudomonas aeruginosa collected in European countries: INFORM global surveillance 2012-15. J Antimicrob Chemother. 2018;73(10):2777–2781.
- Hu YY, Gu DX, Cai JC, et al. Emergence of KPC-2-producing Pseudomonas aeruginosa sequence type 463 isolates in Hangzhou, China. Antimicrob Agents Chemother. 2015;59(5):2914–2917.