ABSTRACT
Introduction: Drug resistance to echinocandins, first-line drugs used to treat Candida auris infection, is rapidly emerging. However, the accumulation of mutations in genes other than FKS1 (before an isolate develops to resistance via FKS1 mutations), remains poorly understood. Methods: Four clinical cases and 29 isolates associated with the incremental process of echinocandin resistance were collected and analyzed using antifungal drug susceptibility testing and genome sequencing to assess the evolution of echinocandin resistance. Findings: Six echinocandin minimum inhibitory concentration (MIC)-elevated C. auris strains and seven resistant strains were isolated from the urinary system of patients receiving echinocandin treatment. Meanwhile, phylogenetic analyses illustrated that the echinocandin-resistant strains were closely related to other strains in the same patient. Genomic data revealed that the echinocandin-resistant strains had FKS1 mutations. Furthermore, three categories (ECN-S/E/R) of non-synonymous mutant SNP genes (such as RBR3, IFF6, MKC1, MPH1, RAD2, and MYO1) in C. auris appeared to be associated with the three-stage-evolutionary model of echinocandin resistance in C. glabrata: cell wall stress, drug adaptation, and genetic escape (FKS mutation). Interpretation: Echinocandin-resistant C. auris undergoes spatial and temporal phase changes closely related to echinocandin exposure, particularly in the urinary system. These findings suggest that FKS1 mutations mediate an evolutionary accumulation of echinocandin resistance followed by modulation of chromosome remodelling and DNA repair processes that ultimately lead to FKS1 hot spot mutations and the development of drug resistance. This study provides an in-depth exploration of the molecular pathways involved in the evolution of Candida auris echinocandin resistance.
Introduction
Candida auris is classified as a critical priority pathogen by the World Health Organization (WHO). Studies indicate that 90% of C. auris isolates are highly resistant to fluconazole, 30% are resistant to polyene amphotericin B, and a small number (<5%) are resistant to echinocandins [Citation1–3]. While echinocandins are the first-line drugs used to treat C. auris infections because of their safety, efficacy, and low resistance rates [Citation4], C. auris-specific echinocandin resistance continues to emerge [Citation5–7]. From 2020 to 2022, we monitored the prevalence of echinocandin resistance in C. auris clinical isolates. Several clinical isolates had elevated minimum inhibitory concentrations (MICs) or even exhibited resistance to micafungin (MCF), caspofungin (CAS), and anidulafungin (ANF).
Research on the mechanism of echinocandin resistance in C. auris has primarily focused on mutations in the FKS1 gene [Citation8,Citation9] (mainly FKS1S639Y, FKS1S639F, and FKS1S639P), but few studies have assessed the drivers of these FKS1 mutations [Citation10]. Importantly, the evolution of echinocandin resistance in multidrug-resistant Candida glabrata strains has multiple steps. It remains unclear whether the development of C. auris-specific echinocandin resistance involves a similar multistep process.
This study focused on a series of C. auris isolates in which resistance emerged following echinocandin treatment. A potential correlation between echinocandin exposure and the emergence of drug resistance was explored. The genome-wide micro evolutionary characteristics of C. auris isolates were analyzed to obtain mechanistic insight into echinocandin resistance. The findings provide a theoretical basis for the prevention, control, and treatment of echinocandin-resistant C. auris infections.
Methods
C. auris strains
The drug sensitivity of C. auris clinical isolates to echinocandins was monitored from 1 December 2020, to 31 July 2022. Patients with elevated echinocandin MICs and echinocandin-resistant strains were enrolled in the study. To further investigate the evolutionary characteristics of C. auris echinocandin resistance, echinocandin-sensitive C. auris strains from these same patients, along with five C. auris isolates (15 September 2022) from the skin and hospital environments of these patients were included in the analysis. These isolates were confirmed as C. auris using MALDI-TOF mass spectrometry.
Clinical data and ethical statement
The medical histories of the enrolled patients with C. auris infection or colonization, including clinical data such as gender, age, department, underlying disease, and antifungal drug use, were obtained from the Hospital Information System (HIS) system of the First Hospital of China Medical University, and assessed by clinicians. This study was reviewed and approved by the Ethics Review Committee of the First Hospital of China Medical University (ERC No. 2024-40) in accordance with the principles of the Declaration of Helsinki.
Drug susceptibility testing
Antifungal susceptibility testing was performed using a commercial chromogenic susceptibility plate (Sensititre YeastOne, Thermo Fisher Scientific). C. auris isolates with MICs ≥4 µg/mL for ANF and MCF, or ≥2 µg/mL for CAS were categorized as echinocandin-resistant (ECN-R) based on the provisional MIC breakpoints published by the Centers for Disease Control and Prevention (CDC) or derived epidemiological cut-off values (ECVs) for C. auris [Citation11,Citation12]. According to Kordalewska et al., MCF is the most potent echinocandin in MIC testing [Citation8]. Thus, C. auris isolates with MCF MICs of 0.5 or 1 µg/mL were categorized as elevated echinocandin MIC (ECN-E) in this study.
Whole genome sequencing
Whole-genome sequencing (WGS) was performed by Shanghai Personal Biotechnology Co., Ltd. (China) using the Illumina NovaSeq platform [Citation3]. Variants that were predicted to alter protein sequences in any coding sequence (non-synonymous single nucleotide variants (SNVs), stop loss or gain variants, indels) were annotated using ANNOVAR software [Citation13] and the RefSeq C. auris B11221 coding sequence.
The cleaned BAM datasets were used to identify copy number variation (CNVs) for each isolate. CNVpytor 1.3.1 was used to identify the genomic regions with CNVs based on the RD analysis approach developed using CNVnator. In brief, this included the following steps: extracting the read depth signal, binning the read depth signal with 100-bp non-overlapping windows across the genome, correcting the signal for GC bias, and segmenting the signal using the mean-shift technique. CNV results with normalized read depth values >1.8 and <0.05 were retained and visualized using R script.
Phylogenetic trees, heat maps, Sankey diagrams, and protein–protein interaction networks
The maximum likelihood method based on the Tamura-Nei model was used to infer evolutionary history [Citation14]. Evolutionary analysis was performed with MEGA7 [Citation15]. A phylogenetic tree was constructed using iTOL software (https://itol.embl.de/). Sanghi diagrams were generated using the SankeyMATIC tool (https://www.chiplot.online/). GenesCloud (https://www.genescloud.cn/) was used to generate heat maps. Protein–protein interaction (PPI) network analysis of non-mutated genes was conducted using the STRING database (https://cn.string-db.org).
Results
Clinical characteristics and drug sensitivity analysis
A total of 29 strains were assessed in this study, including 24 clinical isolates. Detailed strain information is shown in Supplementary 1-1. Seven ECN-R and six ECN-E isolates were identified. Of these, Y0021 was resistant to fluconazole, amphotericin B, and all three echinocandins. Moreover, the echinocandin MICs of in urine specimen isolates from SICU8 increased after treatment. The ANF, CAS, and MCF MICs in C. auris A411 were 0.25, 0.5, and 0.12 µg/mL before treatment, respectively. After two rounds of treatment, all three echinocandin MICs in the A458 (27 June 2022) and A466 (9 July 2022) isolates were 0.5 and 8 µg/mL, respectively. However, SICU8 sputum isolates did not show elevated MICs. The ANF, CAS, and MCF MICs in isolate A427 (26 May 2022) were 0.12, 0.25, and 0.25 µg/mL, respectively, while the MICs of each drug in isolate A459 (27 June 2022) were the same. The specific drug sensitivity results are shown in and Supplementary 1-1.
Table 1. Antifungal susceptibility data for 29 Candida auris.
Case series data for C. auris infection or colonization
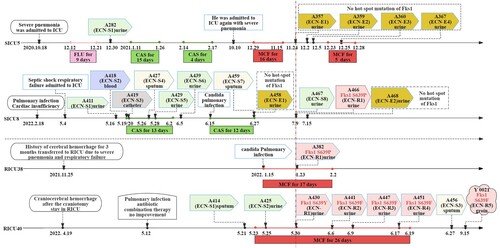
The strains discussed above were isolated from four patients (SICU5, SICU8, RICU38, and RICU40). While SICU5 and SICU8 were admitted to the Surgery Intensive Care Unit ICU (SICU) ward, RICU38 and RICU40 were seen in the Respiratory ICU (RICU) ward. The first echinocandin-sensitive (ECN-S) strain was isolated from case SICU5 at the end of 2020, while the ECN-E strains A357, A359, A360, and A367 were isolated after 1 year of echinocandin treatment. In 2022, ECN-R strains were isolated from cases SICU8, RICU38, and RICU40. The first ECN-E strain from case SICU8, A458, was isolated after the second CAS treatment, and the ECN-R strain, A466, was isolated 12 days later. The ECN-R strain from case RICU38, A382, was isolated after 8 days of MCF treatment, while the ECN-R strain from case RICU40, A430, was isolated after 7 days of MCF treatment. This was followed by the emergence of resistant strains, A441, A447, and A451. All ECN-E and ECN-R strains were isolated from urine samples, while the ECN-S strains were isolated from blood and sputum. Specific case information is shown in and Supplementary 1-2.
Figure 1. Microbiological data and antifungal treatment schedule of the four study patients, showing different intervals of the C. auris strains and echinocandin treatment. Abbreviations ECN-S: echinocandin-sensitive strain; ECN-E: elevated echinocandin MIC strain; ECN-R: echinocandin-resistant strain; CFG: Caspofungin, 50 mg, qd, iv; MFG: Micafugin, 150 mg, qd, iv; FLU: Fluconazole, 200 mg, qd, iv.
Genome sequencing and phylogenetic analysis
Consistent with previous phylogenetic and population structure analyses, all isolates in this study belonged to the South African C. auris clade [Citation3]. A single nucleotide polymorphism (SNP) phylogenetic tree was constructed by combining the 29 C. auris isolates from this study with another 29 isolates from our previous study, including 28 strains from five echinocandin-treated patients and one isolated from the hospital environment (Supplementary 1-3). Of those isolates obtained from our prior study, 25 of 29 were clustered in clade A. Meanwhile, the 29 isolates isolated in this study formed clade B (including B1, B2, and B3 subclades; (A)). Clade B also included four isolates (RICU4 A7, RICU4 A8, RICU9 A52, and C12 A109) from our previous study that may be genetically related to the 29 strains in this study. In addition, all isolates from case RICU40 including his skin strains (Y0021, Y0024), and the Y0006 and Y0010 isolates from a neighbouring patient, case RICU37, were in the B1 subclade and showed a close genetic association. Isolates from SICU5 and his skin strain (Y0013) were in the B2 subclade, while isolates from SICU8 clustered in the B3 subclade. Phylogenetic analysis based on the B1, B2, and B3 subclades suggested that C. auris isolates from the same ward and the same patient were most closely related.
Figure 2. Phylogenetic analysis of the C. auris clinical isolates. (A) Molecular phylogenetic analysis and other relevant information. Phylogenetic trees were generated using the maximum likelihood method of MEGA7 and phylogenetic trees were constructed and composed using iTOL software. All 58 C. auris isolates were obtained from our hospital, half during the present study and half during the previous study. Each sample corresponds to labelled wards, source, date, MICs of MCF, CAS, and ANF, as well as FKS1 mutation site and echinocandin susceptibility status. Abbreviations ECN-S: echinocandin-sensitive strain; ECN-E: elevated echinocandin MIC strain; ECN-R: echinocandin-resistant strain. ECN-R strains and their sample information are marked in red. (B) Statistical plot of the 58 isolates included in the phylogenetic analysis by type, with nine ECN-R strains (red; two from a previous study), six ECN-E strains (yellow), and the remaining ECN-S strains (green). Abbreviations st: stool, ca: catheter, en: environment, bl: blood, sp: sputum, ur: urine.
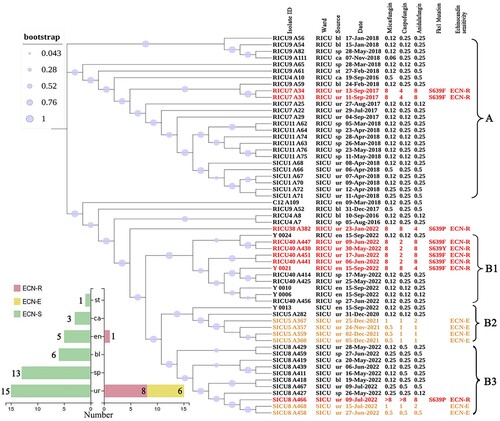
The ECN-R strains were most closely related to other strains in the same patient. Seven ECN-R strains in this study harboured FKS1 mutations (i.e. S639P in A382 and A466, S639Y in A430, and S639F in A441, A447, A451, and Y0021). In addition, eight of the nine ECN-R strains were obtained from urine specimens (two of which were derived from a previous study), and one (Y0021) was derived from the environment (the patient's groin). Six other urine isolates (A357, A359, A360, A367, A458, and A468) were ECN-E ((B)).
Genome-wide SNP locus analysis
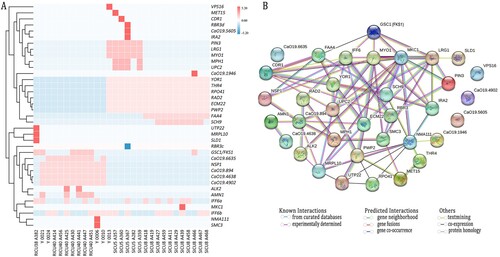
WGS analysis was performed on 29 strains from the C. auris series, of which 47 mutated genes were identified. A total of 35 genes had non-synonymous mutations ((A)), of which 30 had SNPs, five had multiple nucleotide polymorphisms (MNPs), IFF6 (M175I, Y49F), RBR3 (G1385D, S80F), NMA111 (L314F, L811H), AMN1 (C282Y, S123X), FKS1 (S639P, S639F, S639Y), and 12 had synonymous mutations. Almost all the genes had mutations at 1–2 loci of RBR3 and IFF6, but the mutation rate was generally low (i.e. 0.14–0.43), and the non-synonymous mutation rate of the other genes was close or equal to 100%. In addition, the SNP non-synonymous mutant genes in this study did fall into the CNV region, except for one gene, RBR3 (Supplementary Table 1). Of the non-synonymously mutated genes, IFF6 was mutated in all strains, and RBR3 was mutated in most strains. Isolates from RICU40 and RICU37 had seven mutated genes in common, and four urinary isolates from RICU40 (A425, A441, A447, and A451) along with his groin strain (Y0021) had a mutation in AMN1. There were 23 non-synonymously mutated genes in SICU5 and SICU8, of which eight were co-mutated. In addition, SICU5 had 10 unique mutated genes, of which MPH1 and UPC2 were only mutated in ECN-E strains, while SICU8 had five unique mutated genes, of which FKS1 and CaO19.1946 were only mutated in the ECN-R strain, A466. Protein interactions were assessed in 35 non-synonymously mutated genes, of which 31 had interactions. The association analysis of LRG1, MKC1 and GSC1/FKS1 was experimentally determined, text mining and co-expression, The association analysis of MYO1, IFF6, LRG1, MKC1 and GSC1/FKS1 was experimentally determined and text mining, while the association analysis of FAA4 and FKS1 was only co-expression ((B)).
Figure 3. (A): Heatmap of the 29 C. auris strains isolated in this study plotted against their 35 non-synonymously mutated genes using heatmap tools in the genescloud platform (https://www.genescloud.cn). The data are normalized by z-scores. Except for two SNPs in the NMA111 gene of strain Y0006, one SNP appeared in all other genes (shown in red). IFF6 has two mutation sites and is named IFF6a and IFF6b. RBR3 has two mutation sites and is named RBRc and RBRd. (B) Amino acid sequences encoded by 31 non-synonymously mutated genes were used in protein-protein interaction network analysis. Candida albicans SC5314 homologous protein was used as a reference. The identity ranges from 84.5% (GSC1) to 24% (CDR1). Each node in the figure represents a protein, and the edges between the circle nodes represents the interaction relationship between the two proteins linked by the line. Different colours correspond to different interaction types.
Traceability analysis of the characterized genes
Traceability of strain information and plotting of non-synonymously mutated genes of known function in the series of strains from four cases showed that FKS1, ALR2, SLD1, UTP22, and MRPL10 mutations only occurred in the ECN-R strains (). Mutations in MET15, MPH1, CDR1, UPC2, and IRA2 appeared in ECN-E strains while mutations in RBR3, IFF6, YOR1, THR4, PRO41, RAD2, ECM22, PWP2, MYO1, LRG1, PIN3, MKC1, FAA4, SCH9, NSP1, and AMN1 were present in both ECN-S and ECN-R strains.
Figure 4. Sankey diagram showing the summary analysis of genes from case to isolate to MCF MIC and then to non-synonymous mutant SNPS. Two cases (SICU5, SICU8) were SICU ward patients (grey) and two cases (RICU38, RICU40) were RICU ward patients (dark red). Eighteen clinical strains (urine) and one groin specimen, Y0021, were isolated from these four cases. According to the MCF MIC values, a MIC = 0.12/0.25 is regarded as ECN-S (green), MIC = 0.5/1 is regarded as ECN-E (yellow), and MIC = 8 is regarded as drug resistant (red). The non-synonymous mutant SNP gene is present in the strains from three categories (ECN-S/E/R). In addition, the multistep evolution model of C. glabrata echinocandin resistance is associated.
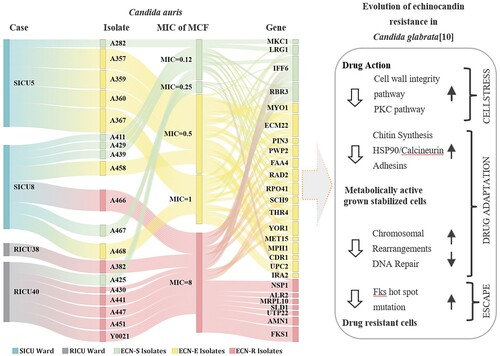
IFF6 and RBR3 were mutated in almost all the early isolates (MIC = 0.12 µg/mL) from the four patients as well as their subsequent isolates. In SICU patients, genes that were co-mutated in cases SICU8 and SICU5, YOR1 and THR4, appeared in case SICU5 strain, A282 (MIC = 0.12 µg/mL), which was collected on 31 December 2020, and were also present in the four ECN-E strains collected from the same patient in 2021 and all isolates collected from case SICU8 in 2022. Early isolate, A282 (MIC = 0.12 µg/mL), from case SICU5 had mutations in MYO1, LRG1, and PIN3, early isolate, A441 (MIC = 0.12 µg/mL), from case SICU8 had mutations in MKC1, FAA4 and SCH9, and early isolate, A425 (MIC = 0.12 µg/mL), from case RICU40 had an NSP1 mutation. MET15, MPH1, CDR1, and UPC2 mutations occurred earliest in caseSICU5 strains with MIC = 0.5 µg/mL (A357, A359, A360), and the IRA2 mutation occurred earliest in case SICU5 strains with MIC = 1 µg/mL (A367). The seven isolates with FKS1 mutations, SICU8 A466, RICU40 A441, RICU40 A447, RICU40 A451, RICU40 A430, RICU38 A382, and Y0021, were resistant to MCF (MIC = 8 µg/mL). In addition to FKS1, RICU38 isolate A382 (MIC = 8 µg/mL) had mutations in the UTP22, SLD1, and MRPL10 genes. The AMN1 mutation, which occurred only in urinary isolates from case RICU40 and its inguinal strain, appeared in the susceptible strain A425 (MIC = 0.12 µg/mL) and its four subsequent ECN-R strains, A430, A441, A447, A451 and Y0021 (MIC = 8 µg/mL).
Three categories (ECN-S/E/R) of the non-synonymous mutant SNP gene in C. auris appears to be associated with the three-stage evolutionary model of echinocandin resistance in C. glabrata: cell wall stress, drug adaptation, and genetic escape (FKS mutation) [Citation10]. Prior research indicates that the non-synonymous mutant genes, RBR3, IFF6, MKC1, and LRG1,which show morphological changes such as cell aggregation in response to drug exposure and other stressors, may be involved at the cell wall stress stage (cell wall integrity or PKC pathways) [Citation16–22]. Meanwhile, RBR3 and IFF6 encode GPI-anchored proteins, which are important cell wall proteins [Citation23]. The MKC1 gene encodes mitogen-activated protein (MAP) kinase, a key component of the PKC pathway [Citation24]. The LRG1 gene encodes Lrg1p, which inhibits the GTPase, Rho1, an activator of the PKC1 pathway [Citation25]. In vivo drug resistance may also involve host environmental stressors [Citation16–22]. The non-synonymous mutant genes, MYO1, MPH1, and RAD2 may be involved in the drug-adaptation stage of echinocandins. The MYO1 gene encodes the heavy chain of myosin [Citation26]. MYO1 is associated with chitin deposition, and MYO1-deficient mutants are sensitive to CAS [Citation27]. MPH1 encodes a DNA fusion enzyme that unwinds the structure of DNA molecules and participates in the DNA repair and recombination process [Citation28]. RAD2 encodes DNA endonuclease, which is involved in nucleotide cleavage and repair [Citation29]. Finally, the genetic escape stage is associated with the FKS1 mutation.
Discussion
The first echinocandin-resistant C. auris was isolated in China (Shenyang) in 2017 [Citation30] and C. auris-specific echinocandin resistance has been closely monitored ever since. This led to the discovery that isolates from four patients, seen from December 2020 to July 2022, had elevated MIC values or echinocandin resistance. Fungal strains from these patients were also monitored at various time points, with particular focus on identifying C. auris clinical isolates before, during, and after echinocandin treatment. Surprisingly, there was a shift from echinocandins-sensitive to drug-resistant strains, indicating that clinical isolates of C. auris in China (Shenyang) were undergoing resistance evolution to echinocandins. Moreover, C. auris (Y0021) isolated from the body surface of RICU40 was resistant to fluconazole, echinocandins, and amphotericin B, indicating a trend toward pan-resistance. These data represent a red flag that requires urgent clinical attention.
Our case series focusing on the evolution of echinocandin resistance in C. auris strains revealed three distinguishing clinical features. First, echinocandin resistance in C. auris is on the rise [Citation3]. Of eight patients monitored during 2016–2020, only one case of echinocandin resistance (RICU7) was noted. However, from the end of 2020 to the end of 2021, ECN-E C. auris strains were isolated from patient SICU5, and from the end of 2021 to June 2022, three patients (RICU38, RICU40, and SICU8) were monitored for the presence of ECN-R strains. Second, the evolution of echinocandin resistance in C. auris strains is closely linked to drug exposure, which is increasing over time. Similar to C. glabrata [Citation10], which is prone to develop multidrug resistance, the four case isolates identified in the present study had elevated MICs or even developed resistance to echinocandins after treatment (). One case, SICU5, was first isolated as a sensitive strain at the end of 2020. After repeated echinocandin exposure over 1 year, SICU5 isolates had a significantly elevated MIC. Subsequently, three cases were identified in 2022 that developed resistance over a significantly shorter period than was seen previously. For example, RICU40 developed echinocandin resistance after 7 days of MCF treatment. In contrast, only one of the eight patient strains in our previous study developed resistance after echinocandin treatment. Based on these results, we hypothesized that after 2022, C. auris acquired a unique potential to develop rapid echinocandin resistance, or even multi-resistance, following treatment. Third, the evolution of resistance to echinocandins in C. auris appeared to originate in the urinary system. Almost all monitored ECN-E and ECN-R strains originated from urine, except for the multi-resistant strain (Y0021), which originated from the inguinal area of RICU40. This may be due to differences in the distribution of echinocandins in each region of the body. To clear infection, high concentrations of echinocandins are concentrated in the blood and sputum [Citation31]. Meanwhile, these drugs are distributed at lower concentrations in the urinary system, which is not sufficient to clear C. auris. In addition, C. auris can colonize the urinary system continuously, and repeated exposure to low drug concentrations may induce resistance. Poor echinocandin penetration in C. glabrata contributes to resistance [Citation32] and suggests that low levels of drug exposure may accumulate additional mechanisms of echinocandin resistance [Citation10]. In agreement with other studies [Citation33,Citation34], the use of echinocandins to treat urinary tract infections or colonization caused by C. auris is not recommended due to the low concentration distribution of echinocandins in the urinary tract. Even when echinocandins are used to treat infections in other regions of the body, close attention should be paid to the development of C. auris echinocandin resistance in the urinary system.
Consistent with prior studies, C. auris echinocandin resistance is directly related to mutations in FKS1 [Citation35]. In this study, FKS1S639P (two strains), FKS1S639Y (one strain), and FKS1S639F (four strains) mutations occurred in seven resistant strains, suggesting that Chinese (Shenyang) strains of C. auris have FKS1 resistance polymorphisms. Of these, the S639F or S639Y mutation was produced only after MCF treatment, while the S639P mutant strains resulted after CAS or MCF treatment. The FKS1 polymorphic variations may be related to exposure to different echinocandin drug types, but further investigation using a larger sample size is required to confirm this.
Most importantly, this study identified the potential drivers of FKS1 mutations in clinical C. auris isolates. Mutated genes from four series of strains that were evolving echinocandin resistance were collected, allowing the identification of 35 non-synonymously mutated genes. Notably, while IFF6 and RBR3 encode cell wall proteins and were commonly mutated in the four series of strains, they were not mutated in any pre-studied strains [Citation3]. This suggests that variation in the IFF6 and RBR3 genes may impact the evolution of C. auris echinocandin resistance. Further study is required to assess this. Six strains of ECN-E C. auris were identified from the SICU ward, and two SICU ward patients (SICU5 and SICU8) had active non-synonymously mutated gene variants, along with the appearance of 23 non-synonymous mutated genes (e.g. MKC1 and MYO1). Given the evolution of C. glabrata resistance to echinocandins [Citation10], MYO1, and other genes may be involved in adaptive changes. Of these, RBR3, IFF6, MKC1, and LRG1, among others, may be involved in cell wall integrity /PKC pathways, MYO1 may be involved in chitin synthesis [Citation26], and MPH1 and RAD2 may be involved in DNA repair and chromatin remodelling [Citation28,Citation29]. These genes may help C. auris to develop echinocandin resistance, allowing the pathogen to tolerate these drugs and provide time for the formation of FKS hotspot mutations needed for drug evasion. Subsequently, six echinocandin-resistant strains with FKS1 mutations were detected in two patients in the RICU wards (RICU38 and RICU40). UTP22, SLD1, MRPL10 and ALK2 were only mutated in the resistant strains, and their involvement in the evolution of echinocandin resistance in C. auris requires further investigation. AMN1 was mutated in five ECN-R strains in RICU40, suggesting that AMN1 may be closely related to the FKS1 mutation in this patient.
The C. auris strains isolated during 2020−2022 were divided into two major evolutionary branches that differed from the isolates obtained before 2020, suggesting that C. auris strains in China (Shenyang) are changing. The C. auris strains isolated in this study were more closely related to blood isolates from the two patients in our previous study, indicating that those C. auris strains that cause bloodstream infections are more likely to disseminate. These findings highlight the importance of strictly controlling the prevalence of C. auris from blood sources to effectively prevent and control widespread dissemination among patients and in the environment. The data also showed that ECN-R strains were more closely related genetically to other isolates from the same patient, suggesting that resistance may evolve independently within individual patients rather than spreading from person to person through a single clone.
This study had some limitations. First, the case number was small, limiting our ability to assess the evolutionary genes and their ability to predict echinocandin resistance. The study findings require validation in follow-up in vitro and in vivo studies. Second, the evolution of C. auris resistance to amphotericin B, which was described in another study, was not explored. Third, the current method (fungus 3) for detecting fungal drug sensitivity does not include echinocandins, which inevitably leads to errors in the detection of echinocandin-resistant strains and underestimates the rate of echinocandin resistance.
In conclusion, there is an urgent need to address the continued emergence of C. auris resistance to echinocandin. Routine and active surveillance should be strengthened, with particular attention paid to C. auris urinary colonization or infection. Echinocandins should be prescribed judiciously to prevent the development of C. auris resistance.
Author contributions
Sufei Tian, Yusheng Wu, Hailong Li, Chen Rong, Na Wu, and Yunzhuo Chu have made substantial contributions to conception and design, or acquisition of data, or analysis and interpretation of data; Ning Jiang and Jingping Zhang have been involved in drafting the manuscript; and Hong Shang has given final approval of the version to be published.
CWS_Editorial_Certificate.pdf
Download PDF (29 KB)Supplementary Table 1.xlsx
Download MS Excel (49.8 KB)Supplementary 1.docx
Download MS Word (32.5 KB)Data availability statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA015722 and CRA015753) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Eyre DW, Sheppard AE, Madder H, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379(14):1322–1331. doi:10.1056/NEJMoa1714373
- Chaabane F, Graf A, Jequier L, et al. Review on antifungal resistance mechanisms in the emerging pathogen Candida auris. Front Microbiol. 2019;10(11):2788. doi:10.3389/fmicb.2019.02788
- Tian S, Bing J, Chu Y, et al. Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: a three-year surveillance study. Emerg Microbes Infect. 2021;10(1):1088–1096. doi:10.1080/22221751.2021.1934557
- Bidaud AL, Chowdhary A, Dannaoui E. Candida auris: an emerging drug resistant yeast – a mini-review. J Mycol Med. 2018;28(3):568–573. doi:10.1016/j.mycmed.2018.06.007
- Sharma D, Paul RA, Rudramurthy SM, et al. Impact of FKS1 genotype on echinocandin in vitro susceptibility in Candida auris and in vivo response in a murine model of infection. Antimicrob Agents Chemother. 2022;66(1):e0165221. doi:10.1128/AAC.01652-21
- Biagi MJ, Wiederhold NP, Gibas C, et al. Development of high-level echinocandin resistance in a patient with recurrent Candida auris candidemia secondary to chronic candiduria. Open Forum Infect Dis. 2019;6(7):ofz262. doi:10.1093/ofid/ofz262
- Jacobs SE, Jacobs JL, Dennis EK, et al. Candida auris pan-drug-resistant to four classes of antifungal agents. Antimicrob Agents Chemother. 2022;66(7):e0005322. doi:10.1128/aac.00053-22
- Kordalewska M, Lee A, Park S, et al. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother. 2018;62(6):e00238-18. doi:10.1128/AAC.00238-18
- Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat. 2007;10(3):121–130. doi:10.1016/j.drup.2007.04.002
- Healey KR, Perlin DS. Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. J Fungi (Basel). 2018;4(3):105. doi:10.3390/jof4030105
- Burrack LS, Todd RT, Soisangwan N, et al. Genomic diversity across Candida auris clinical isolates shapes rapid development of antifungal resistance in vitro and in vivo. mBio. 2022;13(4):e0084222. doi:10.1128/mbio.00842-22
- Arendrup MC, Prakash A, Meletiadis J, et al. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother. 2017;61(6):e00485-17. doi:10.1128/AAC.00485-17
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi:10.1093/nar/gkq603
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi:10.1093/oxfordjournals.molbev.a040023
- Kumar S, Stecher G, Mega TK. Molecular evolutionary genetics analysis. Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi:10.1093/molbev/msw054
- Muñoz JF, Welsh RM, Shea T, et al. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics. 2021;218(1):iyab029. doi:10.1093/genetics/iyab029
- Miramón P, Pountain AW, Lorenz MC. Candida auris-macrophage cellular interactions and transcriptional response. Infect Immun. 2023;91(11):e0027423. doi:10.1128/iai.00274-23
- Zamith-Miranda D, Amatuzzi RF, Munhoz da Rocha IF, et al. Transcriptional and translational landscape of Candida auris in response to caspofungin. Comput Struct Biotechnol J. 2021;19:5264–5277. doi:10.1016/j.csbj.2021.09.007
- Lara-Aguilar V, Rueda C, García-Barbazán I, et al. Adaptation of the emerging pathogenic yeast Candida auris to high caspofungin concentrations correlates with cell wall changes. Virulence. 2021;12(1):1400–1417. doi:10.1080/21505594.2021.1927609
- Shivarathri R, Jenull S, Chauhan M, et al. Comparative transcriptomics reveal possible mechanisms of amphotericin B resistance in Candida auris. Antimicrob Agents Chemother. 2022;66(6):e0227621. doi:10.1128/aac.02276-21
- Shivarathri R, Jenull S, Stoiber A, et al. The two-component response regulator Ssk1 and the mitogen-activated protein kinase Hog1 control antifungal drug resistance and cell wall architecture of Candida auris. mSphere. 2020;5(5):e00973-20. doi:10.1128/mSphere.00973-20
- Bing J, Guan Z, Zheng T, et al. Rapid evolution of an adaptive multicellular morphology of Candida auris during systemic infection. Nat Commun. 2024;15(1):2381. doi:10.1038/s41467-024-46786-8
- Boisramé A, Cornu A, Da Costa G, et al. Unexpected role for a serine/threonine-rich domain in the Candida albicans Iff protein family. Eukaryot Cell. 2011;10(10):1317–1330. doi:10.1128/EC.05044-11
- Wiederhold NP, Kontoyiannis DP, Prince RA, et al. Attenuation of the activity of caspofungin at high concentrations against Candida albicans: possible role of cell wall integrity and calcineurin pathways. Antimicrob Agents Chemother. 2005;49(12):5146–5148. doi:10.1128/AAC.49.12.5146-5148.2005
- Kumari S, Kumar M, Esquivel BD, et al. Unmasking of CgYor1-dependent azole resistance mediated by target of rapamycin (TOR) and calcineurin signaling in Candida glabrata. mBio. 2022;13(1):e0354521. doi:10.1128/mbio.03545-21
- Rodríguez-Quiñones JF, Irizarry RA, Díaz-Blanco NL, et al. Global mRNA expression analysis in myosin II deficient strains of Saccharomyces cerevisiae reveals an impairment of cell integrity functions. BMC Genomics. 2008;9(1):34. doi:10.1186/1471-2164-9-34
- Moreira-Walsh B, Ragsdale A, Lam W, et al. Membrane integrity contributes to resistance of Cryptococcus neoformans to the cell wall inhibitor caspofungin. mSphere. 2022 Aug 31;7(4):e0013422. doi:10.1128/msphere.00134-22
- Marton T, Chauvel M, Feri A, et al. Factors that influence bidirectional long-tract homozygosis due to double-strand break repair in Candida albicans. Genetics. 2021;218(1):iyab028. doi:10.1093/genetics/iyab028
- Sun X, Thrower D, Qiu J, et al. Complementary functions of the Saccharomyces cerevisiae Rad2 family nucleases in Okazaki fragment maturation, mutation avoidance, and chromosome stability. DNA Repair (Amst). 2003;2(8):925–940. doi:10.1016/S1568-7864(03)00093-4
- Tian S, Rong C, Nian H, et al. First cases and risk factors of super yeast Candida auris infection or colonization from Shenyang, China. Emerg Microbes Infect. 2018;7(1):128. doi:10.1038/s41426-018-0131-0
- Howard SJ, Livermore J, Sharp A, et al. Pharmacodynamics of echinocandins against Candida glabrata: requirement for dosage escalation to achieve maximal antifungal activity in neutropenic hosts. Antimicrob Agents Chemother. 2011;55(10):4880–4887. doi:10.1128/AAC.00621-11
- Healey KR, Nagasaki Y, Zimmerman M, et al. The gastrointestinal tract is a major source of echinocandin drug resistance in a murine model of Candida glabrata colonization and systemic dissemination. Antimicrob Agents Chemother. 2017;61(12):e01412-17. doi:10.1128/AAC.01412-17
- Jeffery-Smith A, Taori SK, Schelenz S, et al. Candida auris: a review of the literature. Clin Microbiol Rev. 2018;31(1):e00029-17. doi:10.1128/CMR.00029-17
- Ruiz-Gaitán A, Martínez H, Moret AM, et al. Detection and treatment of Candida auris in an outbreak situation: risk factors for developing colonization and candidemia by this new species in critically ill patients. Expert Rev Anti-Infect Ther. 2019;17(4):295–305. doi:10.1080/14787210.2019.1592675
- Carolus H, Pierson S, Muñoz JF, et al. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. mBio. 2021;12(2):e03333_20. doi:10.1128/mBio.03333-20
