Abstract
Vancomycin resistant enterococci (VRE) are increasingly important causes of morbidity and mortality in developed countries. Although VRE is a significant cause of nosocomial sepsis in these countries, limited data is available on the role that this pathogen plays in South Africa. We describe the demographic, clinical and genotypic data of seven patients involved in the first outbreak of VRE in a haematology unit at a tertiary hospital in Durban and also report the isolation of VRE from six patients from other wards in this hospital and from hospitals outside Durban. The outbreak occurred from 19 April 2011 to 9 November 2011. Pulse Field Gel Electrophoresis (PFGE) was conducted on 15 clinical and environmental samples. Two closely-related clusters and a unique strain were identified from both clinical and environmental samples. Furthermore, the predominant cluster was found in other hospitals in KwaZulu-Natal. After infection control practices were reinforced, the outbreak terminated. Our study highlights that VRE is an emerging pathogen in KZN, especially in high risk units. The environment serves as a significant reservoir of VRE and infection control strategies should be directed to reduce the transmission of VRE from environmental sources.
Introduction
Vancomycin resistant enterococci (VRE) are increasingly important causes of morbidity and mortality in developed countries. In USA, the incidence of VRE infections in 2006 was 6.51 per 10 000 hospital admissions, which had doubled from 2000.Citation1 High prevalence rates have been reported from some European countries where VRE has been found in over 30% of clinical isolates.Citation2 The overall cause and attributable mortality approximates 65% and 40%, respectively, in bacteraemic patients.Citation3−5
Although VRE is a significant cause of nosocomial sepsis in these countries, limited data is available on the role that this pathogen plays in South Africa.Citation6 VRE was present in Cape Town in the early 1990s; however, the first two South African cases were only reported in 1997.Citation7,8 Subsequently, VRE was found as a coloniser in about 11% of Intensive Care Unit (ICU) patients screened at hospitals in Johannesburg.Citation9 Apart from an outbreak of VRE in an oncology unit in Johannesburg in 2000, there has been a paucity of reports of VRE outbreaks from South Africa.Citation10 Furthermore, there are no published reports of VRE from Durban, South Africa. Surveillance data from private and public sector laboratories confirm that there has been an increase in VRE isolated from blood cultures since 2011(C.N. Govind, personal communication, 13 June 2013, and http://www.fidssa.co.za/images/Surveillance_report_NHLS-SASCM_2013).
We describe the demographic, clinical and genotypic data of patients involved in the first reported outbreak of VRE in a haematology unit (HU) at a tertiary hospital in Durban. We also highlight the emergence of VRE as a potential pathogen in KwaZulu-Natal.
Materials and methods
Hospital setting and data collection
The outbreak, which included seven patients over eight months, occurred in the adult HU at Inkosi Albert Luthuli Central Hospital (IALCH), an 846-bed tertiary public sector hospital in Durban. Cases were defined as patients in whom enterococci with a vancomycin minimum inhibitory concentration (MIC) ≥ 32 μg/ml were isolated from any sample, regardless of clinical significance. VRE was first isolated from a urine sample taken on 19 April 2011. The next case of VRE was on 2 August 2011 and there were sporadic cases until the last was identified on 9 November 2011.
In addition, VRE was isolated from two other patients at IALCH, outside the HU as well as from four patients from two private hospitals outside Durban, in the province of KwaZulu-Natal.
Data collected included age, sex, hospital, ward, number of days in the hospital, underlying risk factors, history of diarrhoea and previous antibiotic exposure.
Environmental samples
Samples were taken randomly from rooms in the HU that housed the cases at three different time points when new cases were detected. Seven samples were taken from the room of Patient 2 on 5 August 2011; 50 environmental samples were taken from all the rooms and reception area in the HU on 10 September 2011; and, 45 samples were taken from the five rooms in the HU on 18 October 2011. “High touch” surfaces were sampled, which included drip stands, computer keyboards, mattresses, door knobs, bedrails, tables, commodes and taps.
Microbiological methods
On receipt, all stool, rectal and environmental samples were plated on colistin nalidixic acid agar. Identification and susceptibility testing was performed on colonies resembling enterococci using Vitek 2® system (bioMérieux SA, France). Etests for vancomycin (bioMérieux SA, France) were also performed according to manufacturer’s recommendations.
Isolates were stored at -70 °C until genotyping was performed.
Genotyping
Fifteen isolates from clinical and environmental samples were available for genotyping. Macro-restriction analysis was performed as previously published.Citation9,11,12 Clustering was done according to Tenover criteria. A cluster was defined as Pulsed Field Gel Electrophoresis (PFGE) patterns differing by 3 or less bands.Citation13
Ethics approval was granted by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal (BE230/12).
Results
The outbreak in the HU at IALCH
On 19 April 2011, a vancomycin resistant Enterococcus faecium was isolated in a urine sample from Patient 1 who had been nursed in an isolation room in the general haematology ward prior to the sample being sent. This was the first VRE (all isolates were identified as E. faecium thus VRE signifies vancomycin resistant E. faecium in the context of our study sample) isolated at IALCH.
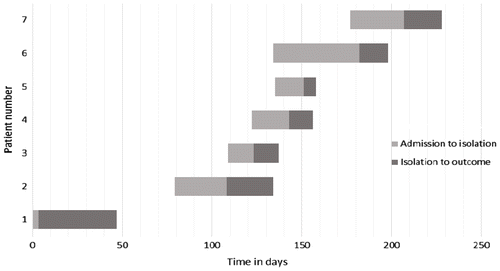
After the second patient with VRE was identified in HU, infection control measures were heightened, which included education of staff, improvement of hand hygiene practices, contact precautions and thorough environmental cleaning. Although isolation and barrier nursing is routinely practiced in the HU, these were reinforced during this period. In addition, all new patients were screened for VRE colonisation using stool or rectal swabs. Patients 5 and 7 were detected during screening. After patient 7, there were no further cases of VRE in the HU. Refer to Figure for the timeline of VRE isolation in HU.
Environmental samples
Two samples from the drip stand and bedside table taken from the room of Patient 2 on 5 August 2011 were positive for VRE. Three samples taken on 10 September 2011 (the door of the toilet in the room of Patient 4 and from commodes in the sluice room) isolated VRE. All 45 samples from five rooms in the HU on 18 October 2011were negative for VRE.
Clinical and demographic data
Vancomycin resistant E. faecium were isolated from 13 patients from 19 April 2011 to 17 February 2012 from Inkosi Albert Luthuli Central Hospital (IALCH), and Lancet Laboratories in Durban. Clinical, demographic and genotypic data are presented in Tables and from IALCH and private patients, respectively. Patients were hospitalised from 3–110 days (median 29 days) before VRE was isolated. The ages of patients ranged from 13–76 years (median 33 years). Nine patients were females and 4 were males. The samples from which VRE was isolated included 4 urine samples, 4 blood cultures, 2 stools, 1 vaginal swab, 1 central venous catheter tip and 1 peritoneal fluid sample. Seven patients had haematological malignancies, one patient had carcinoma of the bladder and one patient had undergone a double valve replacement. Clinical data was unknown for the other patients. Previous antimicrobial exposure, treatment and outcome of all patients are presented in Table . Seven patients were treated for VRE with linezolid; however, two patients demised. In nine patients, data on previous exposure to antimicrobials were available; eight patients received previous broad spectrum antimicrobial therapy, which ranged from 2 -30 days, and five patients had been on vancomycin for 7–20 days (mean 16 days) before VRE was isolated.
Table 1: Clinical and demographic data of 9 patients with VRE at IALCH
Table 2: Clinical and demographic data of 4 patients with VRE at private hosptials in KZN IALCH
Table 3: Antimicrobial exposure and outcome in 13 patients with VRE
Identification and susceptibility test results
All isolates were identified as Enterococcus faecium as they demonstrated the same antibiogram and vanA phenotype. This phenotype was not confirmed genotypically. The MICs generated by the Vitek 2® were as follows: ampicillin ≥ 32 μg/ml, penicillin ≥ 64 μg/ml, vancomycin ≥ 32 μg/ml, teicoplanin ≥ 32 μg/ml, linezolid 2 μg/ml, ciprofloxacin ≥ 8 μg/ml, moxifloxacin ≥ 8 μg/ml, erythromycin ≥ 8 μg/ml, clindamycin ≥ 8 μg/ml, tetracycline ≥ 16 μg/ml, tigecyline ≤ 0.12 μg/ml and trimethoprim-sulphamethoxazole ≥ 320 μg/ml. The isolates also demonstrated a high level resistance to both gentamicin and streptomycin. All isolates demonstrated MICs for vancomycin ≥ 32 μg/ml using Etest™ (Biomerieux, SA).
Genotyping
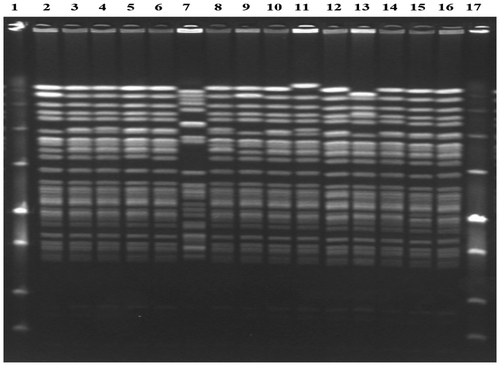
PFGE was conducted on 15 isolates. Tables , and demonstrate the PFGE results of clinical and environmental samples, respectively. Two clusters were identified with isolates in each cluster being indistinguishable. Both clusters were closely related. Cluster A consisted of 10 isolates which were from clinical and environmental samples from the haematology unit, from the peritoneal fluid of a patient in the surgical ICU at IALCH and from three clinical samples from two private hospitals in KwaZulu-Natal.
Table 4: Results of PFGE of five environmental samples from HU
Cluster B consisted of two isolates from a blood culture and a commode in the haematology unit. Cluster A differed by a single band difference from Cluster B and, therefore, they were closely related.
One unique pattern was identified from the blood culture from a patient in the unit who also had an isolate belonging to cluster B from another blood culture taken three days later. Two isolates, closely related to cluster A and B, were from Patient 1 and from an environmental sample in the unit. Figure displays the banding patterns of PFGE for the 15 isolates tested.
Figure 2: PFGE results of all VRE isolates tested.
Discussion
We describe the first outbreak of VRE in KZN which occurred in a haematology unit in Durban. This is the second outbreak of VRE to be reported in South Africa. We also highlight that VRE may be an emerging pathogen in KZN.
Limited data exists on the significance of VRE as a pathogen in South Africa. E. faecium vanB was detected as early as 1993 in Cape Town, followed by four Enterococcus faecalis vanA isolates from Bloemfontein in 1995.Citation7 The first cases reported in 1997, were E. faecalis and E. faecium of the vanA phenotype from Johannesburg, isolated from a blood culture and pus swab from patients previously treated with vancomycin.Citation8 Subsequently, there was an outbreak of Enterococcus faecium vanA genotype in an adult oncology unit in Johannesburg.Citation10 Twenty-two of the 31 strains that were cultured were clonally related. After strict infection control measures were undertaken, the outbreak was terminated. These findings are in keeping with our results in which 10 of 15 isolates belonged to a single cluster. A prevalence study based at four hospitals in Johannesburg that screened high risk patients using rectal swabs, found that almost 11% of those screened, harboured VRE.Citation9 These isolates included 3 E. faecium vanA, 10 E. faecium vanB, 6 Enterococcus gallinarium vanC1 and 1 Enterococcus avium vanA genotypes. Interestingly, the authors demonstrated the clonal spread of E. faecium vanA genotype not only within the hospital but probably also between hospitals in Johannesburg.
Macro-restriction analysis of the isolates revealed two major clusters that were closely related. These clusters comprised both clinical and environmental samples and were also found in all three hospitals. This finding is in keeping with the findings of McCarthy et al.Citation10 Other studies of outbreaks also confirm the spread of one major clone; or, spread of closely-related isolates.Citation14−16 In contrast, Almyroudis et al. demonstrated that most patients in a haematology ward were colonised with unique strains and that spread was unlikely to have occurred from a common source.Citation17
Strategies to reduce the nosocomial transmission of VRE recommended by the Centre for Disease Control (CDC, USA) include education of personnel, active surveillance cultures, prudent antibiotic use, contact isolation, hand hygiene and decontamination of environmental surfaces.Citation18
Environmental contamination is a risk factor for acquisition of VRE.Citation19 We found that the same strains were present in both clinical and environmental samples in the HU. Environmental cultures are positive in up to 30% of outbreak investigations and this may serve as a reservoir for transmission.Citation20 Part of the control plan that we employed at the time of the outbreak was to meet with housekeeping personnel to discuss the importance of environmental cleaning in the control of VRE. Studies have highlighted the value of this strategy by demonstrating a reduction in VRE incidence in ICUs after this was instituted.Citation21,22
Another valuable approach employed in the control of the studied outbreak was the use of active surveillance of cultures and isolation of infected patients. This is supported by a study that showed an eightfold decrease in the incidence of VRE bloodstream infections in a haematology unit using active screening of stool.Citation23 Furthermore, using this policy, mathematical modelling predicted a 39% decrease in annual incidence of VRE in a 10-bed ICU.Citation24
The evidence linking previous antibiotic exposure with resistance is particularly strong for VRE. Apart from previous exposure to vancomycin, other antimicrobials including third-generation cephalosporins, aminoglycosides, ciprofloxacin, meropenem, clindamycin and metronidazole have been independently associated with the VRE carrier state.Citation20,25 In our study, all except one patient had received broad-spectrum antimicrobials and five of these patients had received vancomycin for at least 7 days before VRE was isolated. One of the control strategies for VRE would be to have an effective antimicrobial stewardship programme, which was not in place at the time of the outbreak.
Our study has a few limitations. Samples collected during active surveillance were plated onto colistin nalidixic acid agar plates. Ideally, the samples should have been plated onto selective media containing vancomycin.Citation20 Although active surveillance cultures were recommended for every new admission, some patients were not screened. The vanA phenotype was not confirmed genotypically; although, in the context of a single organism outbreak with high level glycopeptide resistance, it is most probable that these were vanA-carrying isolates. Lastly, PFGE was only conducted on the 15 isolates that were available. Ideally, all isolates should have undergone genotyping.
Our study highlights that VRE is an emerging pathogen in KZN, especially in high risk units and clinicians need to be aware of this. Furthermore, the environment serves as a significant reservoir of VRE and infection control strategies should be directed to reduce the transmission of VRE from environmental sources. Adherence to infection prevention and control policies as well as antimicrobial stewardship programmes should be mandatory in all hospitals in an effort to limit the spread of VRE throughout the province.
Conflict of interest
The authors did/do not have a commercial or other association that might pose a conflict of interest.
Acknowledgments
We would like to thank the patients and staff of the haematology unit and the staff from the microbiology laboratory at Inkosi Albert Luthuli Central Hospital, Durban; Dr A. Niehaus and Dr. A. Singh who collected the environmental samples; and, Dr. P. Mutevedzi who assisted with epidemiological data. Finally, we would like to thank Dr. L. Blann who conducted genotyping of the isolates.
Additional information
Funding
Notes
Presented in part at FIDSAA 10–12 October 2013, Drakensberg.
References
- Ramsey AM, Zilberberg MD. Secular trends of hospitalization with vancomycin‐resistant enterococcus infection in the United States, 2000–2006. Infect Control Hosp Epidemiol. 2009;30:184–6.10.1086/597418
- Werner G, Coque TM, Hammerum AM, et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008;13(47):pii–19046. Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19046.
- Edmond MB, Ober JF, Dawson JD, et al. Vancomycin-resistant enterococcal bacteremia: Natural history and attributable mortality. Clin Infect Dis. 1996;23:1234–9.10.1093/clinids/23.6.1234
- Weinstock DM, Conlon M, Iovino C, et al. Colonization, bloodstream infection, and mortality caused by vancomycin-resistant enterococcus early after allogeneic hematopoietic stem cell transplant. Biol Blood Marrow Transplant. 2007;13:615–21. doi: 10.1016/j.bbmt.2007.01.078.
- Rosa RG, Schwarzbold AV, dos Santos RP, et al. Vancomycin-resistant Enterococcus faecium bacteremia in a tertiary care hospital: epidemiology, antimicrobial susceptibility, and outcome. BioMed Res Int. 2014;2014:958469. doi: 10.1155/2014/958469 . Epub 2014 Mar 5.
- Johnston BL, Bryce E. Hospital infection control strategies for vancomycin resistant enterococcus, methicillin-resistant Staphylococcus aureus and Clostridium difficile. CMAJ. 2009;180:627–31.10.1503/cmaj.080195
- Derby P, Allan B, Lambrick M, et al. Detection of glycopeptide-resistant enterococci using susceptibility testing and PCR. S Afr J Epidemiol Infect. 1998;13:66–9.
- Budavari SM, Saunders GL, Liebowitz LD, et al. Emergence of vancomycin-resistant enterococci in South Africa. S Afr Med J. 1997;87:1557.
- von Gottberg A, van Nierop W, Duse A, et al. Epidemiology of glycopeptide-resistant enterococci colonizing high-risk patients in hospitals in Johannesburg, Republic of South Africa. J Clin Microbiol. 2000;38:905–9.
- McCarthy KM, Nierop W, Duse A, et al. Control of an outbreak of vancomycin-resistant Enterococcus faecium in an oncology ward in South Africa: effective use of limited resources. J Hosp Infect. 2000;44:294–300.10.1053/jhin.1999.0696
- Murray BE, Singh KV, Heath JD, et al. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–63.
- Morrison D, Woodford N, Barrett SP, et al. DNA banding pattern polymorphism in vancomycin-resistant Enterococcus faecium and criteria for defining strains. J Clin Microbiol. 1999;37:1084–91.
- Tenover FC, Arbeit RD, Goering RV, et al. Interpreting Chromosomal DNA Restriction patterns produced by pulsed field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.
- Rupp ME, Marion N, Fey PD, et al. Outbreak of vancomycin‐resistant Enterococcus faecium in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2001;22:301–3.10.1086/iche.2001.22.issue-5
- Ergani-Ozcan A, Naas T, Baysan BO, et al. Nosocomial outbreak of vancomycin-resistant Enterococcus faecium in a paediatric unit at a Turkish university hospital. J Antimicrob Chemoth. 2008;61:1033–9. doi: 10.1093/jac/dkn066.
- Drews SJ, Richardson SE, Wray R, et al. An outbreak of vancomycin-resistant Enterococcus faecium in an acute care pediatric hospital: lessons from environmental screening and a case-control study. Can J Infect Dis Med Microbiol. 2008;19(3):233–6.
- Almyroudis NG, Lesse AJ, Hahn T, et al. Molecular epidemiology and risk factors for colonization by vancomycin-resistant enterococcus in patients with hematologic malignancies. Infect Control Hosp Epidemiol. 2011;32:490–6.10.1086/659408
- Siegel JD, Rhinehart E, Jackson M, et al. Management of multidrug-resistant organisms in health care settings, 2006. Am J Infect Control. 2007;35:S165–S193. doi: 10.1016/j.ajic.2007.10.006.
- Drees M, Snydman DR, Schmid CH, et al. Prior environmental contamination increases the risk of acquisition of vancomycin-resistant enterococci. Clin Infect Dis. 2008;46:678–85. doi: 10.1086/527394.
- Cetinkaya Y, Falk P, Mayhall C. G.. Vancomycin-resistant enterococci. Clin Micro Rev. 2000;13:686–707.10.1128/CMR.13.4.686-707.2000
- Hayden MK, Bonten MJ, Blom DW, et al. Reduction in acquisition of vancomycin-resistant enterococcus after enforcement of routine environmental cleaning measures. Clin Infect Dis. 2006;42:1552–60.10.1086/503845
- Hota B, Blom DW, Lyle EA, et al. Interventional evaluation of environmental contamination by vancomycin-resistant enterococci: failure of personnel, product, or procedure? J Hosp Infect. 2009;71:123–31. doi: 10.1016/j.jhin.2008.10.030.
- Hachem R, Graviss L, Hanna H, et al. Impact of surveillance for vancomycin‐resistant enterococci on controlling a bloodstream outbreak among patients with hematologic malignancy. Infect Control Hosp Epidemiol. 2004;25:391–4.10.1086/iche.2004.25.issue-5
- Perencevich EN, Fisman DN, Lipsitch M, et al. Projected benefits of active surveillance for vancomycin-resistant enterococci in intensive care units. Clin Infect Dis. 2004;38:1108–15. doi: 10.1086/382886.
- Karki S, Houston L, Land G, et al. Prevalence and risk factors for VRE colonisation in a tertiary hospital in Melbourne, Australia: a cross-sectional study. Antimicrob Resist Infect Control. 2012;1:31.10.1186/2047-2994-1-31