Abstract
The study aimed to determine the presence of mumps virus (MuV) in cerebrospinal fluid (CSF) specimens and to genetically characterise detected MuV strains. A real-time reverse transcription-polymerase chain reaction (RT-PCR) was used to detect the MuV F gene, and characterisation was performed by sequencing of the SH gene. Mumps virus was detected in 1.2% (3/260) of specimens. Phylogenetic analysis of one MuV strain revealed that it clustered with the Jeryl-Lynn and RIT4385 vaccine strains. As far as the authors could ascertain this is the first study to provide viral proof that these vaccine-like strains may be associated with aseptic meningitis.
Introduction
Meningitis and encephalitis may complicate an acute mumps virus (MuV) infection, without accompanying parotid swelling in up to 50% of cases.Citation1 Mumps meningitis may, therefore, be clinically indistinguishable from other types of meningitis, and may be underdiagnosed unless specifically tested for. Data on MuV-associated central nervous system (CNS) infections can provide a reflection of the MuV burden in a population.Citation2 Prior to the introduction of the mumps vaccine, MuV was the most common cause of viral meningitis in the United States of America (USA); and, second only to enterovirus as the most common viral cause of meningitis in China.Citation2 The incidence of MuV has since declined in countries with a high mumps vaccine coverage but, in non-vaccinated countries, it is still estimated to be in the order of 100 to 1000 cases per population of 100000.Citation3 The mumps vaccine is not currently included in the South African Extended Program on Immunisation (EPI); and, is only administered in the private health sector. The last epidemiological data regarding MuV meningitis in the mostly unvaccinated population of Gauteng, South Africa dates back 20 years.Citation4
MuV belongs to the Rubulavirus genus within the Paramyxoviridae family. Genotypes are assigned based on sequence analysis of the small hydrophobic (SH) gene, which is the most variable region of the genome.Citation3 It is currently not known which genotype(s) is prevalent in SA.
Currently, all available mumps vaccines contain live attenuated viral strains. Aseptic meningitis may occur after vaccination but, in contrast with wild-type mumps meningitis, no long term sequelae have been reported. The incidence of aseptic meningitis varies between vaccine strains with higher rates reported following Urabe Am9 immunisation, while Jeryl Lynn and Jeryl Lynn-derived strains demonstrate the least neurovirulence.Citation5
The first aim of this study was to determine the presence of MuV in cerebrospinal fluid (CSF) specimens of patients with a clinical presentation suggestive of acute CNS infection. These patients tested negative for other bacterial and fungal pathogens by microbiological culture; and, other clinically suspected viruses by molecular diagnostic testing, conducted in a Gauteng laboratory. The second aim was to characterise detected MuV strains at the molecular level.
Methods
This study was approved by the Faculty of Health Sciences Research Ethics Committee, University of Pretoria, South Africa (Protocol 198/2011). CSF specimens were collected from routine diagnostic specimens received for viral studies in the diagnostic laboratory of National Health Laboratory Service, Tshwane Academic Division, from patients with a clinical diagnosis of meningitis, encephalitis or non-specific febrile illness with or without focal neurological deficit. Specimens that tested positive for the presence of any other infectious agent during routine testing were excluded from the study. Specimens were collected from December 2010 to December 2012 and were stored at -70 °C until use.
Extraction of MuV RNA was done manually using the Nucleospin® RNA XS kit (Macherey-Nagel, Germany). Extracted ribonucleic acid was eluted in 30 μL RNAse free water (Macherey-Nagel, Germany). A previously described one-step real-time reverse transcription-polymerase chain reaction (RT-PCR) assay, which uses primers and probes that detect both the MuV F gene and Homo sapiens RNase gene (RNase P) as an endogenous internal control, was used.Citation6 The 20 μL reaction mixtures were prepared using the QuantiTect RT-PCR kit (Qiagen, Germany). The RT-PCR was performed on the LightCycler® 2.0 platform (Roche Diagnostics, Germany).
A published nested RT-PCR was performed to amplify the entire SH gene (316 base pairs) of MuV strains in MuV-positive specimens.Citation7 The MuV Urabe Am9 vaccine strain was used as a positive control. To reverse transcribe viral RNA to cDNA, 10 μL of RNA was added to an antisense reaction mixture of random hexamers (Roche, Switzerland), nuclease free water (Promega Corp., USA) and 10 mM dNTPs (Promega, USA). The nested RT-PCR reactions were prepared with the EmeraldAmp Max HS PCR Master Mix (Takara Bio, Japan) according to manufacturer’s instructions. The RT-PCR products were detected using 2% agarose gel electrophoresis, which was run at 100V for 40 minutes. RT-PCR products were recovered with the Zymoclean™ Gel DNA Recovery Kit (Zymo Research Corp., USA) according to the manufacturer’s instructions. Amplicons were purified using Zymo Spin Columns (Zymo Research, USA) according to the manufacturer’s instructions; and, sequenced using the Big Dye Terminator Cycle Sequencing Kit v 1.3 (Applied Biosystems, USA). Basic sequence manipulation, verification and alignment were performed using Sequencher 4.7. Multiple sequence alignments were edited using BioEdit version 7.0.9.0 (6/27/07).Citation8 Phylogenetic trees were constructed with maximum-likelihood methods as implemented in the Mega software version 4,Citation9 using reference strains recommended by the WHO in 2012.Citation3
Results
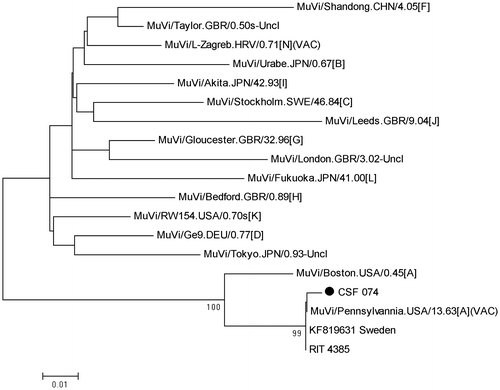
A total of 260 cerebrospinal fluid specimens were selected and screened for the presence of MuV by real-time RT-PCR. MuV was detected in 1.2% (3/260) of specimens. Nucleotide sequence analysis was successful for one of the MuV strains (CSF074). The phylogenetic tree (Figure ) revealed that the MuV strain clustered with the Jeryl-Lynn vaccine strain MuVi/Pennsylvannia.USA/13.63[A](VAC) and the RIT4385 vaccine strain with 99% nucleotide similarity. Further analyses on the other MuV positive specimens were not possible due to insufficient specimen.
Figure 1: Neighbour-joining phylogenetic tree of the South African mumps virus specimen CSF074 in relation to selected other mumps virus nucleotide sequences in GenBank.
Discussion
This study reports a low detection rate of MuV in CSF specimens in the study population. A higher detection rate of 6 (6.7%) MuV from 90 patients with aseptic meningitis was reported in a previous study conducted between 1994 and 1995 in Pretoria, Gauteng, South Africa.Citation4 An older study on CSF of 11 360 aseptic meningitis cases in Cape Town, Western Cape, South Africa, reported a detection rate of 9% of MuV.Citation10 Globally, the proportion of aseptic meningitis attributable to MuV varies depending on vaccination coverage.Citation2 In a recent study in Malawi, where mumps vaccine coverage is reported to be poor, 5.7% of children with suspected viral CNS infection tested positive for MuV.Citation11
Mumps meningitis is a relatively benign condition. As such, some patients may not seek medical attention; and, those who do present to primary or secondary health care centres may not require referral to tertiary care institutions.Citation1 This may account for the lower than expected positivity rate in this study since the majority of the study population were patients hospitalised in a tertiary referral hospital. Furthermore, the specimens were collected over a period of two years and, due to the paucity of surveillance data on MuV in SA, it is not known whether the collection of specimens co-incided with an epidemic peak in MuV activity, which usually occurs every 2 to 5 years.Citation2 The findings may also reflect a lower incidence of MuV-associated CNS manifestations.
Genetic analysis of one detected MuV strain revealed that it has a genotype A Jeryl-Lynn or RIT4385 vaccine-like strain. This is the first study to the authors’ knowledge to provide virological proof that the Jeryl-Lynn MuV vaccine strain may be associated with aseptic meningitis. Post-vaccine aseptic meningitis has been well described following immunisation with mumps vaccines, particularly with the Urabe, Leningrad-3 and Leningrad-Zagreb strains.Citation5 The Jeryl-Lynn strain and its derived RIT4385 strain are considered to be the least neurovirulent of all the MuV vaccine strains.Citation1 Previously described cases of Jeryl-Lynn- and RIT4385-associated aseptic meningitis were presumptive based either on clinical case definitions, such as symptom onset within a specified period after vaccination, or isolation of MuV from CSF with no distinction made between wild type virus and the vaccine strain.Citation5
This study is subject to a number of limitations. This was a retrospective study which may not reflect future epidemiological trends of MuV infection. As mumps aseptic meningitis is usually a benign condition, not all patients will require hospitalisation. The study population may not be representative of MuV infection in the greater society as the majority of specimens were collected from hospitalised patients in a tertiary referral centre. Specimens that tested positive for other pathogens were excluded from the study; therefore, coinfections with MuV would not have been detected, which may have contributed to the lower incidence reported in this sudy. The underlying sero-incidence of MuV in the study population is not known. No vaccination history was available for the patient whose CSF specimen showed 99% nucleotide similarity to the Jeryl-Lynn and RIT4385 vaccine strains.
In conclusion, the current findings suggest that the incidence of MuV-associated CNS disease may be lower in Gauteng and surrounding areas than previously described. The reported MuV vaccine-like strain in CSF highlights the need for MuV surveillance and, specifically, molecular epidemiological studies in partially vaccinated populations. This data can provide a reflection on the MuV-associated CNS infection status and MuV burden in a population. Continued efforts to detect and characterise MuV is, therefore, recommended.
Conflict of interest
The authors declare that they have no conflict of interest.
Declarations
The information contained in this article was presented by Marieke Brauer at the UNIPATH 2014 conference, held on 19-21 September 2014 at the CSIR Convention Centre in Pretoria (no abstract number available).
Additional information
Funding
References
- Hviid A, Rubin S, Mühlemann K. Mumps. Lancet. 2008;371:932–44.10.1016/S0140-6736(08)60419-5
- Galazka AM, Robertson SE, Kraigher A. Mumps and mumps vaccine: a global review. Bull World Health Organ. 1999;77:3–14.
- World Health Organisation. Mumps virus nomenclature update: 2012. Wkly Epidemiol Rec. 2012;87:217–24.
- Engelbrecht L, Brisley T, Taylor MB. Laboratory-based active surveillance of viral aseptic meningitis in Pretoria, 1994-1995. S Afr J Epidemiol Infect. 1996;11:68–71.
- Bonnet MC, Dutta A, Weinberger C, et al. Mumps vaccine virus strains and aseptic meningitis. Vaccine. 2006;24:7037–45.10.1016/j.vaccine.2006.06.049
- Leblanc JJ, Pettipas JP, Davidson RJ, et al. Detection of mumps virus RNA by real-time one-step reverse transcriptase PCR using the Lightcycler platform. J Clin Microbiol. 2008;46:4049–51.10.1128/JCM.01446-08
- Boga JA, De Oña M, Fernández-Verdugo A, et al. Molecular identification of two genotypes of mumps virus causing two regional outbreaks in Asturias. Spain. J Clin Virol. 2008;42:425–8.10.1016/j.jcv.2008.03.010
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
- Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular evolutionary genetics analysis (MEGA) Software version 4.0. Mol Biol Evol. 2007;24:1596–99.10.1093/molbev/msm092
- McIntyre JP, Keen GA. Laboratory surveillance of viral meningitis by examination of cerebrospinal fluid in Cape Town, 1981–9. Epidemiol Infect. 1993;111:357–71.10.1017/S095026880005706X
- Mallewa M, Vallely P, Faragher B, et al. Viral CNS infections in children from a malaria-endemic area of Malawi: a prospective cohort study. Lancet Global Health. 2013;1:e153–60.10.1016/S2214-109X(13)70060-3
