ABSTRACT
Hepatocellular carcinoma (HCC) is a universal problem. The risk factors for HCC could either be environmental or host-genetic-related. The polymorphism of both FOXP3 (rs3761548) and BRAF (V600E) genotypes and alleles was studied in 104 Egyptian HCC patients and 90 healthy subjects. There is a significant increase in the frequency of AC genotype in HCC group, while there is a significant decrease in the frequency of AA genotype in the patient’s group, compared to controls. The CC genotype is completely absent in the control group, while its frequency is 2 (1.92%) in the patient’s group. The C allele frequency is significantly increased in HCC patients (93 = 44.7%) while it is 39 (22.4%) in the control group. However, there is a significant decrease in the A allele frequency in HCC patients (115 = 55.3%) in comparison with controls (135 = 77.6%). The M allele in the BRAF gene (V600E) is significantly increased in HCC patients (90 = 43.69%) while it is 41 (22.78%) in controls. However, there is a significant decrease in the N allele frequency in HCC patients (116 = 56.31%) compared to controls (139 = 77.22%). FOXP3 (rs3761548) and BRAF (V600E) gene polymorphisms could be considered independent risk factors for HCC incidence.
Introduction
Tumorigenesis is a multistep process that drives normal cells to the progressive acquisition of neoplastic features. The progression of cancer is stimulated by the accumulation of genetic defects that alter gene expression that keeps cellular homeostasis. Since 1970, Human cancers have been identified as genetic diseases, so huge efforts have been made to define the genetic alterations implicated in its pathogenesis. These efforts may help in early diagnosis, development of various therapeutic strategies, and prevention of the disease [Citation1].
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and the third leading cause of cancer-related deaths in adults worldwide. It is closely linked to chronic viral hepatitis infections (e.g. hepatitis B and/or C). Other risk factors include direct exposure to alcohol, aflatoxin, and pyrrolizidine alkaloids. Patients having diseases like hemochromatosis, alpha 1-antitrypsin deficiency, metabolic syndrome, and nonalcoholic steatohepatitis (NASH) are at high risk of developing HCC [Citation2]. Globally, men are more likely to develop HCC than women where the literature shows that for every woman, an average of 2.8 men are diagnosed with HCC [Citation3]. The prognosis and treatment of HCC vary according to the tumor histology, size, how far the cancer has spread, and overall health. A late diagnosis of HCC makes its treatment and recovery much more difficult [Citation4]. Researchers try to find new biomarkers to early detect HCC, in a way that may help reduce its incidence [Citation5]. Therefore, this work is devoted to analyzing some genes that may be related to HCC incidences.
FOXP3 (forkhead box P3), DIETER, AIID, JM2, XPID, PIDX, IPEX or scurfin, is an important protein involved in the response of the immune system [Citation6]. This crucial regulatory gene, which affects the growth and operation of regulatory T cells (Treg cells), is found on the short arm of the X chromosome (Xp11.23) and is a member of the forkhead/winged-helix transcription factor family. Numerous researches have explored the frequency of FOXP3 polymorphisms and their relations to autoimmune disorders and malignancies, especially the potentially functional polymorphisms that are situated in the promoter region of FOXP3 that might affect FOXP3 expression. One of these polymorphisms is rs3761548 (C/A) located in the promoter region of FOXP3 [Citation7].
In cancer, the increase in the activity of regulatory T cells may prevent the immune system from destroying cancer cells. In autoimmune diseases, the decrease of regulatory T-cell activity allows other autoimmune cells to attack the body’s own tissues [Citation8–11]. FOXP3 gene can play a crucial role either in inflammation or chronic infections such as hepatitis B, which is considered an important risk factor for carcinoma [Citation12].
The protein belonging to the rapidly accelerated fibrosarcoma (RAF) family of serine/threonine protein kinases is encoded by the BRAF gene (BRAF-1 or BRAF1), located on chromosome 7 (7q34) [Citation13]. This protein has a key role in regulating the MAP kinase/ERK signaling pathway [Citation14]. This pathway is essential for cell division, differentiation, and secretion. The most common mutation in the BRAF gene is the V600E mutation (rs113488022), which is the most frequently identified cancer-causing mutation in melanoma and other cancers including non-Hodgkin lymphoma, colorectal cancer, thyroid carcinoma, and adenocarcinoma of the lung. Any change in the BRAF gene can increase cancer growth and spread. The alteration in the BRAF gene could be a potential therapeutic target rather than one of the key points in HCC carcinogenesis [Citation15].
Subjects and methods
Subjects and blood sampling
A hundred and four patients with HCC and ninety healthy (referred to throughout this text as the control/HCC-free) subjects were included in the study. Samples were collected from patients admitted to the outpatient clinics of the Oncology Department, Faculty of Medicine, Mansoura University, Egypt during the period from June 2022 to Feb 2023. Their ages ranged from 49 to 60 years. The diagnostic imaging of HCC patients including multiphase magnetic resonance imaging, and/or computerized tomography was executed based on the updated guidelines of the 2018 American Association for the Study of Liver Diseases. Exclusion criteria included HCC patients with any type of malignancies, hepatic dysfunctions, renal disorders, autoimmune diseases, obesity complications, alcohol consumption, or endocrine syndromes. The demographic data was obtained from medical archives including age, gender (male/female), and smoking status (positive/negative).
A blood sample of five ml was drawn from each participating subject. This sample was divided into two portions, of which (3 ml) were poured immediately into a dry tube and left for 10 to 15 minutes at room temperature till clotting. Then, it was centrifuged for 10 mins at 2,058 ×g to obtain serum for estimating some biochemical parameters. The samples are stored at −20°C. The genomic DNA was extracted from the other two ml, which was transferred to an EDTA-coated tube for the analysis of FOXP3 (rs3761548) and BRAF (V600E) polymorphisms. The purified genomic DNA from the whole blood of all participants was extracted using the spin column‐based nucleic acid purification Mini kit based on the manufacturer’s guidelines of the GeneJET Whole Blood Genomic DNA (Catalog No.: #K0781; Thermo Fischer Scientific). The kit utilizes silica-based membrane technology in the form of a convenient spin column, Samples were digested with Proteinase K in the supplied lysis solution. The lysate was then mixed with ethanol and loaded onto the purification column, where the DNA binds to the silica membrane. Impurities are effectively removed by washing the column with the prepared wash buffers. Genomic DNA was then eluted under low ionic strength conditions with the elution buffer [Citation16]. The concentration and absorbance of extracted genomic DNA were evaluated with the aid of the full spectrum NanoDropTM ND‐1000 Spectrophotometer.
Ethics approval and consent to participate
The Institutional Review Board of the Faculty of Science (IRBFS) approved the current study under the code number (SCI-CH-p-2021–196). Written consent was obtained from the legal guardians of each participant. Their data have been declared private and confidential. This study was performed in accordance with the ‘Declaration of Helsinki in 1964’.
Biochemical analysis
Test kits from Biodiagnostic, Egypt, were utilized for the estimation of liver and kidney function. Assays included measuring serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), and serum creatinine.
Genetic analysis
Genotyping of FOXP3 rs3761548 gene polymorphism
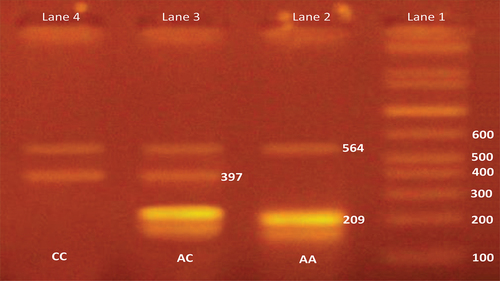
Primers for FOXP3 rs3761548 genotyping were as following: OF: 5′-GACTTAACCAGACAGCGTAG-3′, IF: 5′-TTCTGGCTCTCTCCCCAACTGC-3′ (G allele specific), IR: 5′-TGAGGGGTAAACTGAGGCCTT-3′ (A allele specific), OR: 5′-CTGGTGTGCCTTTGGTCT-3′. Polymerase chain reaction (PCR) was carried out in a final volume of 20 μl reaction mixture containing 2.5 μl of 100 ng DNA, 2.5 μl Mgcl2, 1.25 μl of Taq DNA polymerase and 1 μl of each primer, 2.5 μl dNTPs, and 1 μl PCR buffer. PCR conditions were initial denaturation at 94°C for 5 min followed by 29 cycles of 94°C for 30 s, annealing at 53.5°C and extension at 72°C for 30 s. The final extension was at 72°C for 5 min and holds at 4°C. The amplified products were run on 2% agarose gel containing ethidium bromide at 100 V for 20 min. The A allele specific product showed a band at 209 base pair (bp) and C allele specific band at 397 bp with general product at 564 bp [Citation17], see .
Genotyping of BRAF (V600E) gene polymorphism
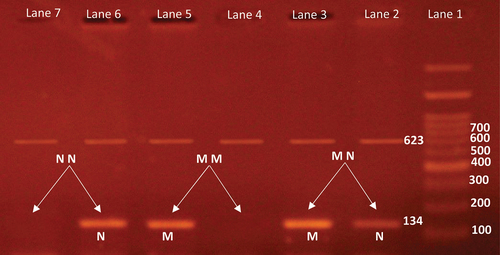
A forward and two reverse primers were used in the PCR reaction. The forward primer was 5’-GCTTGCTCT- GATAGGAAAATGAG-3“, while the two reverse primers were 5”-ACCCACTCCATCGAGATTTCT-3’ (mutation-specific) and 5’-CTGTGGATCACACCTGCCTTA-3’ (control). The 25 μl reaction mixture consisted of 1.25 U Taq polymerase, 160 μM dNTPs, 1X PCR Buffer, 1.5 mM MgCl2 and 100 ng genomic DNA. PCR was carried out with the initial denaturation for 15 min at 95°C. It consisted of 40 cycles. Each cycle was 40 s at 94°C, 40 s at 61°C, and 40 s at 72°C. Final extension for 5 min at 72°C. The general band appeared at 623 bp, while the mutation band was 134 bp, see . All PCR products were separated on 2% agarose gel after staining with ethidium bromide [Citation18].
Figure 2. Agarose gel electrophoresis of BRAF(V600E) gene.
Statistical analysis
The raw data have been analyzed using the JMP software, version 16. Qualitative data are presented as frequencies. However, quantitative normally distributed data are presented as mean ± standard deviation (SD). Deviations from Hardy-Wein-Berg equilibrium expectations were determined using the goodness of fit between the observed and expected genotype frequencies. HWE indicates that the selected groups of study are reasonable for performing genetic analysis of this single nucleotide polymorphism (SNP). Kolmogorov – Smirnov test was done to test the normality of data distribution. Skewed distributed data have been presented as median (lower quartile – upper quartile). Significance was accepted at p < 0.05.
Results
The demographic and clinical characteristics of both HCC cases and healthy volunteers are shown in . Our analyzed results show a highly significant increase in the levels of liver enzymes, creatinine, and alpha-fetoprotein (AFP) in the HCC group in comparison with their levels in the control group (p < 0.001**). It is worth mentioning that 30.4% of our HCC patients’ sample is diabetic, 60% of them have Anti-HCV, while only 5.6% of them have HBs Ag. They are all significantly different (with p-value <0.001) in comparison with the healthy volunteer group.
Table 1. The demographic and clinical characteristics of both HCC cases and healthy volunteers.
shows the frequency distribution of both genotypes and alleles of the genes FOXP3 and BRAF(V600E) in both control and patient groups. The FOXP3 genotypes are AA, AC, and CC. Analyzing the data revealed that there is a highly significant increase in the frequency of the AC genotype in the HCC group when compared to its corresponding in the control group. Their frequencies are respectively 89 (85.58%) and 39 (44.83%). However, there is a significant decrease in the frequency of the AA genotype in the patients’ group in comparison with the control group, which are, respectively, 13 (12.5%) and 48 (55.17%). The CC genotype is completely absent in the control group, while its frequency in the patient’s group is 2 (1.92%). Keeping CC+AC as a baseline, the odds for the AA genotype frequency was calculated at 8.6, with a 95% confidence interval of (4.2–17.7) and with a p-value <0.0001. The C allele frequency is significantly higher in the group of HCC patients (93 (44.7%)) than in the group of HCC-free individuals (39 (22.4%)). However, the frequency of A allele is significantly lower in the HCC group (115 (55.3%) than in the control group (135 (77.6%)). The odds ratio for the allelic frequencies was also significant with a p-value <0.0001, odds ratio of 2.8, and a 95% confidence interval of (1.79–4.39), see .
Table 2. FOXP3 rs3761548 and BRAF (V600E) genes distribution in both HCC cases and healthy volunteers.
The genotypes of the BRAF gene (V600E) are MM, MN, and NN. It is shown in that the frequency of the MN genotype is significantly higher in the HCC group than in the control group. It is 82 (79.61%) and 37 (41.11%) in the HCC and HCC-free groups, respectively. However, the frequency of the NN genotype is significantly lower in the HCC patients’ group (17 (16.5%)) than in the control group (51 (56.67%)). Also, the frequency of the MM genotype is 4 (3.88%) in the control group and 2 (2.22) % in the patients group. The odds for the NN genotype frequency was calculated at 0.15, with a 95% confidence interval of (0.08–0.29) and with a p-value <0.0001, with reference to MM+MN as a baseline. The M allele is significantly increased in the group of HCC patients 90 (43.69%), while it is 41 (22.78%) in the control group. On the contrary, the N allele frequency is significantly decreased in the HCC group (116 (56.31%) when compared with the HCC-free group (139 (77.22%)). The table shows further that the odds ratio for the BRAF gene allelic frequencies was also significant with a p-value <0.0001, odds ratio of 0.38, and a 95% confidence interval of (0.24–0.59).
The genotype frequency distributions of FOXP3 rs3761548 and BRAF (V600E) genes in smokers and nonsmokers having HCC are shown in . The table shows that there is no significant difference in the frequency distribution of both FOXP3 rs3761548 and BRAF (V600E) genotypes and alleles in both groups of smokers and nonsmokers.
Table 3. Genotype frequencies of FOXP3rs3761548 and BRAF gene (V600E) polymorphisms in smokers and nonsmokers individuals having HCC.
The influence of acquiring hepatitis C virus infection on the gene polymorphism of FOXP3 rs3761548 and BRAF (V600E) genes has been analyzed and presented in . The table shows that there is no significant difference in the frequency distribution of both genotypes and alleles neither in FOXP3 rs(3761548) nor in BRAF (V600E) genes in both groups (positive and negative anti-HCV), except in the NN genotype where there is a significant decrease in negative anti-HCV compared to positive- HCV ones; with a p-value = 0.026, odds ratio of 4.14 and a 95% confidence interval of (1.10–15.56).
Table 4. Genotype frequencies of FOXP3 (rs3761548) and BRAF gene (V600E) polymorphisms in positive and negative anti-HCV individuals having HCC.
The potential effects of HCC family history as well as the acquisition of diabetes mellitus on the frequency distribution of FOXP3 rs3761548 and BRAF (V600E) gene polymorphism in HCC patients have been analyzed and shown in , respectively. shows that there is no significant difference in the frequency distribution of both genotypes and alleles neither in FOXP3 nor BRAF in both groups (with and without a family history). A similar result has been obtained for the case of diabetes mellitus, see , where it is shown that the presence or absence of diabetes mellitus accompanied by HCC does not significantly alter the distribution of the genotype frequencies.
Table 5. Frequency distribution analysis of FOXP3 rs3761548 and BRAF (V600E) gene polymorphisms in HCC patients with family history.
Table 6. Genotype frequencies of FOXP3 rs3761548 and BRAF gene (V600E) polymorphisms in diabetic and non-diabetic HCC patients.
In silico data analysis
The bioinformatics frameworks of the gene Fork Head Box Protein 3 (FOXP3) are demonstrated in . The Fork Head Box Protein 3 is positioned on chromosome X and spanned about 14,273 bp [chrX:49,250,438–49,264,710]. [Ensembl; ENSG00000049768]. The cellular FOXP3 localized to the nucleoplasm in addition localized to the cytosol [Data source: Cellular compartment database]. Genetic and protein interaction networks suggested that the FOXP3 is a transcriptional regulator which is essential for the development and inhibitory function of (Treg) cells. It has a vital role in maintaining homeostasis of the immune system. It acts either as a transcriptional repressor or a transcriptional activator depending on its interactions with other transcription factors. The number of transcripts is 7 and it interacts with 11 proteins [Data source: STRING and GeneMania databases].
Figure 3. Genomic structure of the human forkhead box P3 (FOXP3)[Homo sapiens (human) gene (FOXP3)]. a) Subcellular localization of the FOXP3 protein, with darker colors indicating more copiousness. b) 3D structure of the FOXP3 protein. c) FOXP3 gene coexpression. d) Protein – protein interaction using the STRING database. e) Gene-gene interaction using gene MANIA database. f) FOXP3 gene interaction and pathways from curated databases and text-mining, genes in the interaction graph are connected by a number of different line types, with each type of line and the line properties themselves indicating different levels of support from text mining and databases. [data source: Ensembl.Org, NCBI database, compartment database, X2K web database, gene mania, and STRING version 11.0].
![Figure 3. Genomic structure of the human forkhead box P3 (FOXP3)[Homo sapiens (human) gene (FOXP3)]. a) Subcellular localization of the FOXP3 protein, with darker colors indicating more copiousness. b) 3D structure of the FOXP3 protein. c) FOXP3 gene coexpression. d) Protein – protein interaction using the STRING database. e) Gene-gene interaction using gene MANIA database. f) FOXP3 gene interaction and pathways from curated databases and text-mining, genes in the interaction graph are connected by a number of different line types, with each type of line and the line properties themselves indicating different levels of support from text mining and databases. [data source: Ensembl.Org, NCBI database, compartment database, X2K web database, gene mania, and STRING version 11.0].](/cms/asset/11e4b5a7-5385-4e30-9300-b088336da0b7/teba_a_2300209_f0003_oc.jpg)
The bioinformatics frameworks of the gene the human B-Raf proto-oncogene or serine/threonine kinase (BRAF) are demonstrated in The BRAF gene is positioned on chromosome 7 and spanned about 205,593 bp [chr7:140,719,337–140,924,929].
Figure 4. Genomic structure of the human B-Raf proto-oncogene or serine/threonine kinase (BRAF)[Homo sapiens (human) gene (BRAF)]. a) Subcellular localization of the BRAF protein, with darker colors indicating more copiousness. b) 3D structure of the BRAF protein. c) BRAF gene coexpression. d) Protein – protein interaction using the STRING database. e) Gene-gene interaction using gene MANIA database. f) BRAF gene interaction and pathways from curated databases and text-mining, genes in the interaction graph are connected by a number of different line types, with each type of line and the line properties themselves indicating different levels of support from text mining and databases. [data source: Ensembl.Org, NCBI database, compartment database, X2K web database, gene mania, and STRING version 11.0].
![Figure 4. Genomic structure of the human B-Raf proto-oncogene or serine/threonine kinase (BRAF)[Homo sapiens (human) gene (BRAF)]. a) Subcellular localization of the BRAF protein, with darker colors indicating more copiousness. b) 3D structure of the BRAF protein. c) BRAF gene coexpression. d) Protein – protein interaction using the STRING database. e) Gene-gene interaction using gene MANIA database. f) BRAF gene interaction and pathways from curated databases and text-mining, genes in the interaction graph are connected by a number of different line types, with each type of line and the line properties themselves indicating different levels of support from text mining and databases. [data source: Ensembl.Org, NCBI database, compartment database, X2K web database, gene mania, and STRING version 11.0].](/cms/asset/b344d2d9-2e9d-4c31-8795-7be4f06ac8a4/teba_a_2300209_f0004_oc.jpg)
[Ensembl; ENSG0000015776]. The cellular BRAF localized to the vesicles and cytosol [Data source: Cellular compartment database]. Genetic and protein interaction networks suggested that the BRAF protein plays an important role in regulating the MAP kinase/ERK signaling pathway. It affects cell division, differentiation, and secretion. The mutations BRAF gene is most commonly the V600E mutation. This mutation is the most frequently identified cancer-causing mutation in melanoma and in various other cancers. The number of Transcripts is 5. It interacts with 14 proteins. [Data source: STRING and GeneMania databases].
Discussion
Hepatocellular carcinoma is considered a big health problem worldwide. The highest incidence and mortality of HCC are reported in East Asia and Africa [Citation19]. There are many risk factors involved in HCC incidence. The risk factors can be subdivided into environmental-related risk factors and host-genetic-related risk factors [Citation20]. The incidence and mortality of HCC are still ascending due to the abuse of alcohol, and the acquisition of diabetes mellitus, metabolic syndrome, obesity, and cirrhosis [Citation21].
It has been revealed that the aberrant generation of regulatory T cells is caused by the lack of a functioning FOXP3 gene product. It has been shown that FOXP3 is expressed in both Treg and tumor cells of individuals with cancer; the expression level and activity of FOXP3 may represent a novel route of immune evasion by cancer. The inactivation of the tumor suppressor activity of the FOXP3 gene may be involved in the development of cancer. In another previous study, FOXP3 had a comparable immunosuppressive impact on liver cancer [Citation22].
The existence of functional polymorphic domains in the DNA-binding sites of the promoter region of the FOXP3 gene may have an impact on the expression of the FOXP3 gene. Consequently, there may be a disruption in Treg cell activation and development. The rs2232365 A/G and rs3761548 A/C polymorphisms in the FOXP3 gene promoter have been linked to a number of illnesses, including multiple sclerosis, autism, allergic rhinitis, vitiligo, non-small cell lung cancer, and hepatocellular carcinoma [Citation23].
To date, few publications are available regarding HCC and FOXP3 polymorphisms. Thus, our study investigated the genotype distribution of FOXP3 promoter SNP (rs3761548) in Egyptian HCC patients in order to illustrate their implication on the development/prognosis of HCC.
FOXP3 gene has a crucial role in the development and pathogenesis of cancer. FOXP3 is expressed in Treg cells. Therefore, it defines primary Tregs in patients with cancers [Citation24]. Chen et al. [Citation23] concluded that FOXP3 rs3761548 gene polymorphism contributes to the susceptibility of colorectal cancer in the Chinese population. Also, Chen et al. [Citation25] reported that FOXP3 rs3761548 polymorphism is associated with cancer risk. Consequently, the polymorphisms of FOXP3 could not only change its level of expression but also impair the suppressive function of Tregs. Therefore, it gives FOXP3 a vital role in the early detection and elimination of the progression of HCC disease [Citation26]. Chen et al. concluded that FOXP3 rs3761548 polymorphism may play a principal role as a vital biomarker to predict oral cancer disease development and prognosis [Citation27].
The meta-analysis of Chen et al. analyzed three types of FOXP3 polymorphisms and six types of cancers. The analysis revealed that FOXP3 rs3761548 polymorphism was associated with increased cancer risk in the Chinese population while FOXP3 rs2280883 and rs 3,761,549 polymorphisms were not [Citation28]. Our results showed that the FOXP3 rs3761548 gene significantly varied in both genotypes and alleles in the selected HCC group compared with the control subjects. Therefore, the FOXP3 rs3761548 gene polymorphism could be considered an independent risk factor for HCC incidence and may help to give new strategies for HCC treatment.
According to demographics and etiologies associated with HCC, many signaling pathways are specifically affected. Tumor suppressor gene (TP53) mutations are the most prevalent alterations in nations with high aflatoxin exposure and long-term HBV carriage. However, given the variability in HCC development, additional routes might be also involved. According to studies conducted in Asian, Italian, and American populations, other RAS/RAF/MAP kinase and PI3K/AKT/mTOR genes were found to be altered. More than half of HCC cases have active MAPK and AKT/mTOR pathways, which are accompanied by proliferative class aggressive tumors. Modifications in the RAS/RAF MAP kinase pathway are progressively being investigated in hepatocellular carcinoma, with potential implications for patient-specific therapy [Citation29].
Many cancer-driver mutations in the onset of HCC are recognized in genes. According to extensive data supplied by the International Cancer Genome Consortium, numerous genes, including the serine/threonine kinase (BRAF) gene and the proto-oncogene B-Raf, are altered in HCC. BRAF is a vital component of the mitogen-activated protein kinases/extracellular signal-regulated kinases (MAPK/ERK) signaling pathway. It is a member of the RAF family of serine/threonine protein kinases and regulates a variety of cellular processes, including apoptosis, stress responses, proliferation, and differentiation [Citation30].
Two decades of research have demonstrated that abnormal signal transduction and modifications in the MAPK/ERK pathway are important initiators of several forms of cancer [Citation31]. Mutations in KRAS and BRAF are linked to about 30% and 8% of all cancer types, respectively [Citation32]. Thymidine (T) to adenosine (A) transversion at nucleotide 1799 is the most prevalent mutation in the BRAF gene, as a consequence, valine (V) is substituted with glutamic acid (E) in codon 600 (V600E) [Citation33]. Mutations in BRAF V600E result in continuous signal transduction and high kinase activity in a RAS-independent manner, which raises the rate of cell division and increases resistance to apoptosis [Citation34]. Additionally, this substitution causes a rise in protein expression or activity that may disrupt the MAPK signaling pathway and cause a variety of developmental problems, including several malignancies in humans [Citation35].
The BRAF pathway plays a crucial and vital role in HCC pathogenesis [Citation36]. Japanese and Chinese studies evidenced that BRAF mutations participate in the etiopathogenesis of HCC [Citation37]. Nansy et al. [Citation38] conducted a study on colorectal cancer (CRC) patients and reported that protein expression of BRAF V600E is not correlated with the prognosis of CRC. Souleymane et al. [Citation29] studied BRAF mutations in HCC Senegal patients. The authors demonstrated that BRAF mutations have no role in tumor metastasis. Sun et al. [Citation39] found that the BRAF gene is associated with capsule formation in HCC Chinese patients. Somi et al revealed that this polymorphism might be involved in gastric adenocarcinoma progression by influencing Tregs functions and the secretion of immunomodulatory cytokines [Citation40].
Based on the discovery of mutations in the known cancer driver gene BRAF in a patient with liver cancer for the first time, Smiech et al. established the tumorigenic impact of BRAF V600E on hepatocytes (THLE-2 cell line). They revealed V600E mutations to be pathogenous in HCC. Most significantly, they discovered a number of genes that are disturbed by THLE-2 cells’ BRAF V600E [Citation30].
To the best of our knowledge, no previous studies investigated the association between BRAF (V600E) gene polymorphism and the risk of HCC among Egyptian population. Our results of BRAF (V600E) revealed that both the genotypes and alleles of the gene are significantly different in the selected HCC patients compared to the control group.
Conclusion
Our study evidenced that both FOXP3 rs3761548 and BRAF (V600E) polymorphisms could participate in increasing susceptibility to HCC. Therefore, both FOXP3 rs3761548 and BRAF (V600E) might be considered independent risk factors for HCC incidence. Hence, they could be used as early predictors for HCC incidence and to prevent the progression of the disease. Also, both FOXP3 rs3761548 and BRAF (V600E) polymorphisms could be used as new biomarkers for the early diagnosis of the disease or as a part of a new strategy in HCC treatment. Besides, future studies might be performed to evaluate the role of these two polymorphisms and their association with the risk of other types of cancer.
Abbreviations
| AFP | = | alpha-fetoprotein |
| ALT | = | Alanine aminotransferase |
| Anti-HCV | = | Hepatitis C virus antibodies |
| AST | = | Aspartate aminotransferase |
| CI | = | Confidence internal |
| FOXP3 | = | forkhead box P3 |
| HBs Ag | = | Hepatitis-B virus surface antigen |
| HCC | = | Hepatocellular carcinoma |
| IQR | = | Interquartile range |
| MAPK/ERK | = | Mitogen-activated protein kinases/extracellular signal-regulated kinases |
| NASH | = | Nonalcoholic steatohepatitis |
| OR | = | Odds ratio |
| PCR | = | Polymerase chain reaction |
| RAF | = | rapidly accelerated fibrosarcoma |
| SNP | = | Single nucleotide polymorphism |
| Treg cells | = | regulatory T cells |
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Naro C, Cunliffe HE, Sette C. Editorial: insight in cancer genetics: 2022. Front Oncol. 2022;12. doi: 10.3389/fonc.2022.988310
- Caruso S, O’Brien DR, Cleary SP, et al. Genetics of hepatocellular carcinoma: approaches to explore molecular diversity. Hepatology. 2021;73(S1):14–26.. doi: 10.1002/hep.31394
- Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–491. doi: 10.1053/j.gastro.2018.08.065
- Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1–61..
- Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7(3):308–319. doi: 10.1016/j.gendis.2020.01.014
- Fondevila F, Fernández-Palanca P, Méndez-Blanco C, et al. Association of FOXO3 expression with tumor pathogenesis, prognosis and clinicopathological features in hepatocellular carcinoma: a systematic review with meta-analysis. Cancers (Basel). 2021;13(21):5349.. doi: 10.3390/cancers13215349
- You D, Wang Y, Zhang Y, et al. Association of Foxp3 promoter polymorphisms with susceptibility to endometrial cancer in the Chinese Han women. Medicine. 2018;97(18):e0582. doi: 10.1097/MD.0000000000010582
- Loo CS, Gatchalian J, Liang Y, et al. A genome-wide CRISPR screen reveals a role for the non-canonical nucleosome-remodeling BAF complex in Foxp3 expression and regulatory T cell function. Immunity. 2020;53(1):143–157. doi: 10.1016/j.immuni.2020.06.011
- Motawi TMK, Sadik NAH, Sabry D, et al. rs62139665 polymorphism in the promoter region of EpCAM is associated with hepatitis C virus-related hepatocellular carcinoma risk in Egyptians. Front Oncol. 2022;11:754104. doi: 10.3389/fonc.2021.754104
- Motawi TMK, Sadik NAH, Sabry D, et al. rs2267531, a promoter SNP within glypican-3 gene in the X chromosome, is associated with hepatocellular carcinoma in Egyptians. Sci Rep. 2019;9(1):6868. doi: 10.1038/s41598-019-43376-3
- Saleh R, Elkord E. FoxP3+ T regulatory cells in cancer: prognostic biomarkers and therapeutic targets. Cancer Lett. 2020;490:174–185. doi: 10.1016/j.canlet.2020.07.022
- Beyzaei Z, Shojazadeh A, Geramizadeh B. The role of regulatory T cells in liver transplantation. Transpl Immunol. 2022;70:101512.. doi: 10.1016/j.trim.2021.101512
- Malapelle U, Rossi G, Pisapia P, et al. BRAF as a positive predictive biomarker: focus on lung cancer and melanoma patients. Crit Rev Oncol Hematol. 2020;156:103118. doi: 10.1016/j.critrevonc.2020.103118
- Barreno LRQ, Mello JBHD, Barros-Filho MC, et al. Characterization of BRAF mutation in patients older than 45 years with well-differentiated thyroid carcinoma. Braz J Otorhinolaryngol. 2022;88(4):523–528. doi: 10.1016/j.bjorl.2020.07.007
- Saad AM, Abdel‐Megied AE, Elbaz RA, et al. Genetic variants of APEX1 p. Asp148Glu and XRCC1 p. Gln399Arg with the susceptibility of hepatocellular carcinoma. J med virol. 2021;93(11):6278–6291. doi: 10.1002/jmv.27217
- Lopez-Martin JA, Fernández AA, Ríos-Martín JJ, et al. Frequency and clinicopathological profile associated with braf mutations in patients with advanced melanoma in Spain. Transl Oncol. 2020;13(6):100750. doi: 10.1016/j.tranon.2020.100750
- Khan MA, Massey S, Ahmad I, et al. FOXO1 gene downregulation and promoter methylation exhibits significant correlation with clinical parameters in Indian breast cancer patients. Front Genet. 2022;13:842943.. doi: 10.3389/fgene.2022.842943
- Machnicki MM, Glodkowska-Mrowka E, Lewandowski T, et al. ARMS-PCR for detection of BRAF V600E hotspot mutation in comparison with real-time PCR-based techniques. Acta Biochimica Polonica. 2013;60(1). doi: 10.18388/abp.2013_1951
- Llovet JM, De Baere T, Kulik L, et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18(5):293–313. doi: 10.1038/s41575-020-00395-0
- Rashed WM, Kandeil MAM, Mahmoud MO, et al. Hepatocellular Carcinoma (HCC) in Egypt: a comprehensive overview. J Egypt Natl Canc Inst. 2020;32(1):1–11. doi: 10.1186/s43046-020-0016-x
- Koulouris A, Tsagkaris C, Spyrou V, et al. Hepatocellular carcinoma: an overview of the changing landscape of treatment options. J Hepatocell Carcinoma. 2021;8:387. doi: 10.2147/JHC.S300182
- Talaat RM, Seada SM, Mohamed GI, et al. Polymorphism of FOXP3 in Egyptian patients with hepatocellular carcinoma (HCC). EC Gastroenterol Dig Syst. 2018;5:97–106.
- Akgöllü E. Evaluation of forkhead box P3 gene polymorphisms in chronic HBV infection. J Gene Med. 2020;22(6):e3172.. doi: 10.1002/jgm.3172
- Chen J, Rong X, Liu X, et al. FOXC2 is a prognostic biomarker and contributes to the growth and invasion of human hepatocellular carcinoma. Cancer Cell Int. 2020;20(1):1–13.. doi: 10.1186/s12935-020-01265-0
- Motawi TMK, Sabry D, Shehata NI, et al. Impact of FOXP1 rs2687201 genetic variant on the susceptibility to HCV‐related hepatocellular carcinoma in Egyptians. J Biochem & Molecular Tox. 2022;36(3):e22965.. doi: 10.1002/jbt.22965
- Cheng Z, Guo Y, Ming L. Functional Foxp3 polymorphisms and the susceptibility to cancer: an update meta-analysis. Medicine. 2018;97(34):e11927. doi: 10.1097/MD.0000000000011927
- Chen PJ, Lin CW, Lu HJ, et al. The impact of FOXP3 polymorphisms on oral cancer progression and clinicopathological characteristics. J Cancer. 2023;14(7):1195. doi: 10.7150/jca.84470
- Chen Y, Qi X, Bian C, et al. The association of FOXP3 gene polymorphisms with cancer susceptibility: a comprehensive systemic review and meta-analysis. Biosci Rep. 2019;39(3). doi: 10.1042/BSR20181809
- Souleymane T, Thorpe DFKS, Mawuli SD, et al. KRAS and BRAF mutations in patients with hepatocellular carcinoma in Senegal. IJGG. 2022;10(1):37. doi: 10.11648/j.ijgg.20221001.16
- Śmiech M, Leszczyński P, Wardell C, et al. Oncogenic mutation BRAF V600E changes phenotypic behavior of THLE-2 liver cells through alteration of gene expression. Int J Mol Sci. 2022;23(3):1548.. doi: 10.3390/ijms23031548
- Terrell EM, Durrant DE, Ritt DA, et al. Distinct binding preferences between Ras and Raf family members and the impact on oncogenic Ras signaling. Molecular Cell. 2019;76(6):872–884.. doi: 10.1016/j.molcel.2019.09.004
- Guo YJ, Pan WW, Liu SB, et al. ERK/MAPK signalling pathway and tumorigenesis (review). Exp Ther Med. 2020;19:1997–2007. doi: 10.3892/etm.2020.8454
- Poulikakos PI, Sullivan RJ, Yaeger R. Molecular pathways and mechanisms of BRAF in cancer therapy. Clin Cancer Res. 2022;28(21):4618–4628.. doi: 10.1158/1078-0432.CCR-21-2138
- Loo E, Khalili P, Beuhler K, et al. BRAF V600E mutation across multiple tumor types: correlation between DNA-based sequencing and mutation-specific immunohistochemistry. Appl Immunohistochem Mol Morphol. 2018;26(10):709–713. doi: 10.1097/PAI.0000000000000516
- Tounsi-Guettiti H, Traina H, Ayed IB, et al. BRAF V600E and novel somatic mutations in thyroid cancer of Libyan patients. Asian Pac J Cancer Prev: APJCP. 2022;23(12):4029.. doi: 10.31557/APJCP.2022.23.12.4029
- Gnoni A, Licchetta A, Memeo R, et al. Role of BRAF in hepatocellular carcinoma: a rationale for future targeted cancer therapies. Medicina. 2019;55(12):754. doi: 10.3390/medicina55120754
- Zhou X, Huang JM, Li TM, et al. Clinical significance and potential mechanisms of ATP binding cassette subfamily c genes in hepatocellular carcinoma. Front Genet. 2022;13:805961.. doi: 10.3389/fgene.2022.805961
- Nansy YA, Mona A, Rania AH, et al. Clinicopathological significance of BRAF V600E mutation in Egyptian colorectal cancer patients. Med J Cairo Univ. 2020;88(June):991–997. doi: 10.21608/mjcu.2020.110833
- Sun W, Zhang Y, Liu B, et al. Gene polymorphism of MUC15, MMP14, BRAF, and COL1A1 is associated with capsule formation in hepatocellular carcinoma. Can J Gastroenterol Hepatol. 2021;2021:1–8. doi: 10.1155/2021/9990305
- Ezzeddini R, Somi MH, Taghikhani M, Moaddab SY, Shirazi KM, Shirmohammadi M, … & Farrokhi AS. Association of Foxp3 rs3761548 polymorphism with cytokines concentration in gastric adenocarcinoma patients. Cytokine. 2021;138:155351. doi: 10.1016/j.cyto.2020.155351