ABSTRACT
Synthetic gene drives could provide new solutions to a range of old problems such as controlling vector-borne diseases, agricultural pests and invasive species. In this paper, we outline methods to identify hazards and detect potentially adverse ecological outcomes at the individual (genotype, phenotype), population, community and ecosystem level, when progressing Gene Drive Modified Organisms through a phased test and release pathway. We discuss the strengths and weaknesses of checklists and structured hazard analysis techniques, identify methods to help meet some of the challenges of detecting adverse ecological outcomes in experiments and confined field trials, and discuss ways to improve the efficiency and statistical rigour of post-release monitoring strategies.
Introduction
Gene drive is a generic term for a variety of processes that in sexually reproducing organisms cause genes to be transmitted to successive generations at ratios greater than the classical Mendelian ratio (Champer, Buchman, and Akbari Citation2016). Natural gene drive processes, such as under dominance and Homing Endonuclease Genes (HEGs), have been known for many decades (Zimmering, Sandler, and Nicoletti Citation1970), and identified as mechanisms to suppress or modify populations of disease vectors (Wood and Newton Citation1991; Braig and Yan Citation Citation2002; Burt Citation2003; Gould, Magori, and Huang Citation2006; Lindholm et al. Citation2016). Harnessing natural drive mechanisms to achieve vector control, however, has proven to be difficult (Smidler, Min, and Esvelt Citation2017), but this situation could change due to the development of RNA-guided endonucleases such as CRISPR/Cas9 (Jinek et al. Citation2012).
CRISPR/Cas9 is a gene-editing technique (Le Cong et al. Citation2013) that is easier and cheaper to implement than other gene-editing tools (Gaj, Gersbach, and Barbas Citation2014), and has consequently reduced the time needed to complete gene-editing experiments from years to days (Saey Citation2015). Furthermore, if constructs are developed with DNA sequences that are homologous with the DNA on either side of the cleavage site, then CRISPR/Cas9 systems can be programmed to act like HEGs, copying themselves into cleavage sites via homology-directed repair, and thereby creating a construct that, if copied into germ-line cells, is inherited at a super-Mendelian rate (Gantz and Bier Citation2015).
CRISPR/Cas9 has been used to successfully edit the genomes of a wide variety of organisms, (Sander and Joung Citation2014; Doudna Citation2015; Gantz et al. Citation2015; Hammond et al. Citation2015), and the potential applications of CRISPR/Cas9 gene-drive systems are diverse (Burt Citation2014; Esvelt et al. Citation2014; NASEM Citation2016a; Burt et al. Citation2018; Leitschuh et al. Citation2017; Scott et al. Citation2017). However, the novelty and relatively simplicity of gene editing with CRISPR/Cas9 systems, together with the potential for creating constructs that (at least theoretically) can spread to an entire population following the release of just a single organism (low threshold gene drives), introduces the possibility of unintended consequences to ecological and epidemiological endpoints (Webber, Raghu, and Edwards Citation2015).
Recognition of these potential risks has led to recommendations for researchers to act cautiously and implement multiple, independent confinement strategies to prevent unintended releases into the environment of organisms carrying gene-drive constructs (Araki, Nojima, and Ishii Citation2014; Akbari et al. Citation2015; NASEM Citation2016a). Ultimately, however, many of the potential benefits of gene-driving technologies are only possible if organisms carrying these constructs are intentionally released into the environment in a manner that is designed to allow them to spread through a target population.
Deliberate unconfined releases of gene-drive modified organisms (GDMOs) will likely be sanctioned within a risk-based decision-making process. This process will be influenced by ethical, socio-cultural, epidemiological, ecological and economic considerations, and the risk analysis process should include mechanisms that facilitate the effective engagement of stakeholders and help integrate these considerations within the overall decision-making process (NRC Citation1996; Renn Citation2008; NASEM Citation2016a). Risk analyses for living modified organisms (LMOs) are typically conducted on a case-by-case basis and we envisage the same approach will apply to GDMOs. These analyses should aim to: (1) identify hazards, defined here as the circumstances that may lead to adverse (harmful) outcomes; (2) evaluate the risks associated with these hazards; (3) identify strategies that help to reduce these risks and (4) design and implement monitoring strategies to detect, or confirm the absence of, adverse outcomes.
In this paper, we focus on this first and last step, by outlining methods to identify hazards and subsequently detect, as efficiently as possible, any potentially adverse ecological outcomes associated with the confined and unconfined release of GDMOs. Identifying potentially adverse outcomes, and the causal pathways by which they might manifest, will help analysts quantify risks, implement risk mitigation strategies and design and implement cost-effective monitoring programmes.
Phased test and release pathway
CRISPR/Cas9 gene-drive systems are theoretically reversible, theoretically limitless, systems capable of altering and suppressing (possibly to extinction) populations of target species (Burt Citation2003; Smidler, Min, and Esvelt Citation2017). A prudent way to approach a deliberate release of a GDMO into the environment for such purposes is to follow an iterative, phased, test and release pathway () that: (a) gathers risk-relevant data under controlled, contained conditions, and thereby develops confidence that the GDMO can safely progress to the next phase; (b) generates release-relevant data by observing outcomes under increasingly realistic scales and conditions, which will require a deliberate and gradual relaxation of the level of containment and (c) terminates the development of GDMOs that fail to meet pre-specified (in a Target Product Profile), phase-specific, performance- and safety-related criteria (NASEM Citation2016a), that may be amended in an iterative fashion based on data gathered at subsequent phases.
Figure 1. Example of phased testing and release pathway for Gene Drive Modified Organisms, based on the phased testing pathway for Genetically Modified Mosquitoes (source: World Health Organization Citation2009). Combined function in Phase 1 refers to the phenotypic effect of the transgene once it is successfully integrated into the genome. Confined field trials in Phase 3 can entail one or more types of physical, ecological or molecular containment strategies. Semi-field conditions in the context of genetically modified mosquitoes refers to enclosed greenhouses that emulate natural environments. More generally, however, semi-field conditions refers to contained release strategies under natural or natural-like conditions.
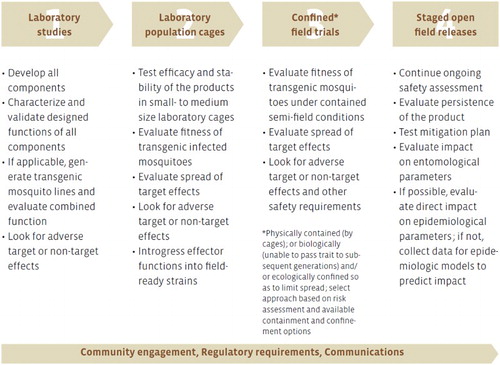
The need to gather risk- and release-relevant data – that is data that helps quantify the likelihood of hazards at increasingly realistic scales – is well understood in the context of transgenic crops (Rissler and Mellon Citation2000; Firbank, Lonsdale, and Poppy Citation2005; Snow et al. Citation2005; Andow and Zwahlen Citation2006). Phased test and release strategies that aim to generate such data have since been recommended for genetically modified mosquitoes (; World Health Organisation Citation2009, Citation2014), transgenic fish (Hayes et al. Citation Citation2014) and most recently GDMOs (NASEM Citation2016a).
Phased test and release strategies for GDMOs should incorporate physical, reproductive, ecological and molecular barriers (Akbari et al. Citation2015). The level of containment provided by physical barriers, such as those described in Arthropod Containment guidelines (ACME/ASTMH Citation2003), will likely vary as a GDMO progresses through the testing and release pathway. We expect physical containment procedures to be most stringent in Phase 1 or Phase 2, possibly exceeding Biosafety Level 2 or Arthropod Containment Level 2 (Benedict et al. Citation2017), because relatively little will be known about the phenotypic characteristics of the GDMO or the reliability of molecular containment strategies in these early phases. If implemented correctly, these procedures are expected to reduce the probability of environmental exposure to very low levels; Hunt and Tabachnick (Citation1996), for example, estimate the probability of small insects (<2 mm in length) escaping their Biosafety Level 3 facilities to be between 1 × 10−10 and 2 × 10−14. These procedures will need to be deliberately relaxed to increase the scale and realism of the experimental environment, if a decision is made to proceed to the next phase.
Ecological barriers – i.e. performing experiments outside the habitable range of the GDMO, or in areas without potential wild mates – is a potentially effective containment strategy. It can be gradually relaxed, by releasing the GDMO to a geographically isolated location within the habitable range, or within a location habitable for only part of the year, and it may eventually be removed entirely. Typically this would occur at the confined field trial or open field release phases of the process. The efficacy of this barrier can be overestimated if our knowledge about the organism’s ability to hybridize with closely related species is incomplete. It can also be circumvented by accidental or deliberate transport to new regions, as demonstrated by the escape of the calicivirus agent for Rabbit Hemorrhagic Disease Virus from field trials on Wardang Island (Mutze, Cooke, and Alexander Citation1995).
Reproductive strategies – i.e. incorporating gene drives in laboratory strains that cannot reproduce with wild organisms – are in principle likely to be highly effective but only suitable for studying drive systems in Phases 1 and 2. Molecular containment methods, and related molecular reversal methods (Esvelt et al. Citation2014; DiCarlo et al. Citation2015; Wu, Luo, and Gao Citation2016), can be theoretically maintained throughout the test and release pathway, but their efficacy in field conditions remains untested.
Identifying hazards
The Royal Society (Citation1983) defines hazard as a situation that in particular circumstances could lead to harm. This definition emphasizes that hazard analysis must identify the circumstances that lead to harm, i.e. the causal pathway, rather than simply identifying potential adverse outcomes. This is useful because the potential adverse ecological outcomes that are being discussed in the context of GDMOs () are the same as, or similar to, those that were identified for transgenic crops (Rissler and Mellon Citation2000; Hilbeck et al. Citation2005; Snow et al. Citation2005; Andow and Zwahlen Citation2006), transgenic fish (Muir and Howard Citation2002; Devlin et al. Citation2007), biocontrol agents and invasive species (Simberloff Citation2012; Simberloff et al. Citation2013). The risks associated with the next generation of GDMOs could be different, however, because the causal pathways that lead to adverse outcomes – and hence the likelihood and magnitude of these outcomes – may be different from those associated with LMOs, biocontrol agent or invasive species.
Table 1. Example hazardous events identified in the literature (second column) that may be relevant to risk analyses of gene drive modified organisms, because of the potentially adverse ecological outcomes (fourth column) that these events might lead to.
The lack of familiarity with a new technology like gene drives presents a challenge to risk analysts who want to identify environmental hazards – i.e. the circumstances leading to adverse ecological outcomes – associated with the deliberate or accidental release of GDMOs. There are two approaches to this challenge: (a) seek precedence in the hazards identified for more or less similar situations, hereafter termed the ‘checklist-like approach’, and (b) employ structured hazard identification to postulate what might go wrong. We explore the strengths and weaknesses of both options below.
Checklist-like approach
Compiling lists of the ways in which adverse outcomes have occurred in the past, and how to prevent similar outcomes in the future, is one way to identify hazards and help ensure that mitigation strategies are applied to the risks of an activity. Checklists are most commonly used in the context of activities that have a long history of successful operation – i.e. where the operating history provides assurance that the risks are well understood and have been managed successfully in the past, e.g. greenhouses or cages in ‘semi-field systems’ (Knols et al. Citation2003; Ferguson et al. Citation2008; for checklist see http://johnmm.bol.ucla.edu/containment.htm#BreachesOfContainment).
A potential drawback of checklists is that hazard identification is focused on what is known to have occurred in the past. A checklist may therefore lead assessors to miss hazards that are unique to a new technology because it operates in ways that are different from existing technologies. Changes in farming practices associated with herbicide tolerant (HT) crops, for example, had positive and negative effects on biodiversity in the UK large-scale field trials (Hawes et al. Citation2003), but virtually all prior commentaries on HT crops failed to identify farming practice change as a potential hazard.
Checklists could also mislead risk analysts into believing that all aspects of a system that ought to have been questioned actually were addressed, or conversely lead analysts to address hazards that are irrelevant to the type of gene drive or its intended application. , for example, provides a list of hazardous events drawn from published studies. This list could serve as a hazard checklist but it is by no means complete, nor necessarily appropriate, because much of the current literature focuses on the use of CRISPR/Cas9 to control mosquito-borne diseases. Thus, analysts seeking to complete a hazard analysis for an RNA-guided Cpf1 endonuclease (Champer, Buchman, and Akbari Citation2016) to suppress mice populations, for example, might miss hazards if they relied solely on . As the range of drive mechanisms and applications grows (NASEM Citation2016a), analysts will need more robust methods to rigorously identify hazards in a comprehensive but case-specific manner.
Structured hazard identification
Structured hazard analysis tools () aim to elucidate the causal pathways that link initiating events at a molecular level to harmful outcomes at an individual, population, community or ecosystem level of organization. Thinking of hazards in terms of causal pathways – or pathways to adverse outcomes – has proven useful in ecotoxicological risk assessments (Ankley et al. Citation2010) but the application of the tools listed here to facilitate this process in ecological risk assessment is still relatively novel.
Table 2. Summary of structured hazard analysis techniques that could be applied to identify hazards associated with GDMOs.
Most of the tools in were developed to identify hazards in industrial/aerospace systems (Kumamoto and Henley Citation1996). Some have been successfully applied to biological systems (Hayes Citation2002a, Citation2002b; Hayes et al. Citation2004), while others were specifically developed for applications in ecological systems (Dambacher, Li, and Rossignol Citation2003). The principal advantage of these methods, as compared to a checklist, is that they help analysts to carefully and systematically apply their expertise beyond their own experience when constructing causal pathways. For example, one possible hazard (out of many) identified in is that the Cas9 protein complex cleaves loci with similar, but not identical, homology to the target loci, leading to the creation of a new phenotype with enhanced capacity to transmit diseases or pathogens. Structured hazard identification tools assist an analyst to construct models of how this hazard could occur, for example by systematically considering the genes that determine or mediate the underlying parameters of disease transmission, the extent to which these genes contain sequences that are homologous to the target loci, and the processes that determine or may alter the sequence specificity of the Cas9 guide RNA. Carefully deconstructing each of the necessary steps in the causal pathway of a particular hazard helps generate risk hypotheses that can be further analyzed and constructively critiqued before proceeding to the next stages of a risk assessment.
In biological settings, the experts needed to complete these processes are usually volunteers drawn from multiple institutions and this can sometimes make it difficult to secure enough time to complete the processes. These processes also require careful facilitation in order to integrate successfully the competencies of groups with often diverse backgrounds and expertise.
Another challenge is that the techniques listed in require a detailed description of the system of interest. At a molecular level this requires knowledge of the biological processes that govern endonuclease promotion, translation, sequence specificity and DNA cleavage and accurate homolog directed repair (e.g. Sander and Joung Citation2014). At the population/community level, they require knowledge of the target organisms’ life-cycle, phenotype, habitat preferences and trophic interactions at each life-stage (e.g. David et al. Citation2013; Hayes et al. Citation Citation2014). The analyst must be able to describe the system, as completely as possible, at an appropriate scale and degree of resolution, in the same way that an engineer would describe an industrial system when implementing these techniques in an industrial/aerospace setting. For biological systems this is a difficult task, which leaves open the possibility that some pathways will be missed.
Identification and detection within the phased testing pathway
Phase 1 and 2: laboratory studies and small-scale experiments
Potential hazards, and their associated adverse outcomes, can be modelled and searched for experimentally (phenotype, population and community outcomes) or via genome sequencing (genetic outcomes). As a GDMO progresses through the phased testing and release pathway, however, the spatial and temporal scales of the concomitant biosafety studies increase, and the suite of tools used to identify hazards and detect adverse outcomes changes ().
Figure 2. Spatial and temporal scales of outcomes associated with the phased test and release pathway of Gene Drive Modified Organisms. As a GDMO progresses through the test and release pathway (phases 1 to 4 in yellow) the spatial and temporal scale of experimental outcomes, and potentially adverse outcomes, increases. The techniques used to identify hazards and detect potential adverse ecological outcomes vary as these spatio-temporal scales change.
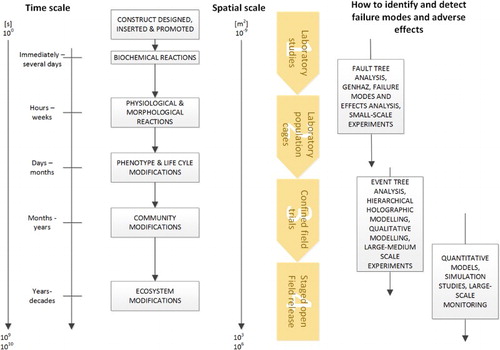
Experimental studies of GDMOs in phases 1 and 2 (respectively, laboratory studies and laboratory population cages in ) are anticipated to follow a three-step process: (i) describe the experimental objectives within the context of the GDMO’s planned genotypic and phenotypic and characteristics (Target Product Profile); (ii) select observational targets from a range of potential targets (within available resources) and (iii) develop and execute an adequate (for the objectives) experimental design.
The experimental objectives in Phase 2 would typically include measuring the gene drive’s conversion efficiency (the rate at which heterozygous individuals are converted to a homozygous state) within a wild-type genetic background. The objectives should also include gathering data relevant to the likelihood of potentially adverse outcomes identified by the hazard analysis methods described previously. For example, the hazard analysis might identify the possibility of off-target changes to near homologous sequences in the organism’s genome that are known or thought to govern traits that influence the GDMO’s environmental niche, its susceptibility to existing control methods or its capacity to transmit animal or plant pathogens. This would indicate the need to conduct genetic screening and/or whole-of-organism experiments to test the frequency of these outcomes. The key challenge, however, is that some of the causal pathways identified by the hazard analysis may have a very low expected frequency, or a very high natural variability, thus making experimentation laborious, prohibitively expensive or simply impossible given the size and generation time of the organism concerned.
There are some model organisms that can be raised in very large (109–1012) numbers and have very short generation times, such as yeast (Botstein, Chervitz, and Cherry Citation1997) or the nematode Caenorhabditis elegans. With these organisms, it is theoretically possible to raise the experimental sample size to a sufficiently large number such that ‘rare’, but important events, like spontaneous genetic mutations that compromise molecular containment systems (Moe-Behrens, Davis, and Haynes Citation2013) can be detected. All such experiments, however, come with the caveats that the evolutionary dynamics of gene drives within model organisms can only approximate those of other species, and cannot account for possible gene by environment (Tabachnick, Citation2003; Lobo Citation2008) or epigenetic effects that may occur within real environments.
If the likelihood of a hazard that leads to an undesirable genetic or phenotypic outcome in a target organism is extremely rare, it may be possible to identify influential, intermediate, events on the causal pathway that are expected to occur at a higher frequency, using for example Fault Tree Analysis (FTA), and associated sensitivity analysis techniques (Iman Citation1987; Rausand and Hoyland Citation2004; Aven and Nøkland Citation2010; Hayes et al. Citation2015). If experimental evidence can determine the rate of one or more intermediate events then, contingent on the structure of the fault tree (including the extent to which it is expanded or pruned) and the assumption that the tree is an adequate model, this may provide enough evidence for decision-making purposes that the fault tree’s top event will be sufficiently unlikely. For example, researchers may not have to complete an exhaustive search for near homology across all possible genes that govern an organism’s capacity to transmit disease (the top event) if the probability of an essential intermediate event on the causal pathway, such as a change in the timing or manner in which the cas gene is expressed (the intermediate event), was quantified and shown to be sufficiently low.
In this context, it is also not necessary for the intermediate event in question (or even a top event) to be actually observed in an experiment. Null event inference methods (for the so-called zero numerator problem) can be used to establish confidence or credible intervals for the probability of events that are not observed in independent experiment trials (Wilson Citation1927; Winkler, Smith, and Fryback Citation2002; Gerlach, Mengersen, and Tuyl Citation2009). So long as the experimental trials are independent these methods can be used to estimate the probability of the event in question, which may be sufficient for decision-making purposes. In this context, however, identifying independent trials may not be straightforward. For example, in an experiment that crosses wild-type organisms with a GDMO to test for undesired phenotypes, it may not be immediately clear what constitutes an independent trial – the number of zygotes, the number of mating events or the number of GDMOs used in the experiment.
Phase 3: confined field trials
The phased testing and release pathway deliberately seeks to increase the realism of experiments by moving outside of the laboratory confines. This is likely to be important for several reasons. Firstly, experience with Bt crops (Lövei and Arpaia Citation2004; Duan et al. Citation2009) and biocontrol agents (Barratt et al. Citation2010) suggests that the relevance of laboratory experiments for some types of hazards, such as non-target effects and host specificity, can be equivocal and therefore contested.
Secondly, the phenotype of a transgenic organism could be influenced by environmental conditions and the organism’s genetic background (Tabachnick, Citation2003). Genetically modified salmon, for example, can be much larger than wild conspecifics when raised in standard hatchery conditions, but only slightly larger when raised under naturalized stream conditions (Sundström et al. Citation2007). Phase 3 provides an opportunity to examine these (gene by environment) effects by testing how the GDMO performs relative to the criteria established in its Target Product Profile, under natural or semi-natural field conditions, whilst still maintaining a relatively high degree of containment (compared to Phase 4).
This phase may also enable analysts to design experiments targeting population- and community-scale hazards that arise through causal pathways that involve changes to, or interactions with, wild-type organisms (Pennington et al. Citation2010) and a realistic suite of predator/prey organisms. Knols et al. (Citation2003), for example, describe semi-natural environments, including breeding sites, feeding sites, indigenous plants and real predators built within contained greenhouses to conduct experiments with the Anopheles gambiae mosquito complex.
Whilst Phase 3 offers a number of advantages over Phase 2, research in Phase 3 will likely face a number of challenges. The probability of inadvertent escape and spread may be higher (depending on the type containment strategy used) than Phase 2. Effect size may be much smaller (as in the case of Sundström et al. Citation2007), replication could be more expensive, and outcomes may be more variable than those observed under laboratories conditions (Facchinelli et al. Citation2013). It may therefore be difficult to achieve a satisfactory degree of statistical power. Finally, semi-natural field experiments may fail to adequately capture the conditions that the GDMO experiences during an open field release, and may therefore show outcomes that do not occur in the field (Nguyen et al. Citation2015).
Phase 4: open field release
As the evaluation of a GDMO moves to Phase 4 (open field release in ), the task of designing experiments or monitoring programmes to detect potentially adverse outcomes, and attributing them to the GDMO if they are detected, becomes more difficult. The same basic three-step process described under Phases 1 and 2 still applies but some elaboration is required to deal with two additional challenges:
The increased number of possible confounding effects outside of a controlled environment.
The increase in scope from mainly temporal variability within the small spatial footprint of contained field trials, to the logistically more challenging and potentially more heterogeneous, spatial and temporal scope of the uncontained field release.
Deliberate release strategies will be able to choose release sites, but will likely have limited control over the subsequent spread of a GDMO within the boundaries set by ecological and reproductive barriers. This means it may be more difficult to identify and/or maintain appropriate control sites.
Data collected in the field are typically more representative of the population of interest than data collected during controlled (e.g. randomized) experimental trials because the latter are often conducted in constrained environments (Rubin Citation1974). The difficulty with field data, however, is that the analyst may not be able to adequately account for the effect of confounding variables, particularly in the absence of an extensive pre-release monitoring programme that helps establish a comparative baseline. Experience with the yield of Bt crops (NASEM Citation2016b) and the effect of biocontrol agents (Barratt et al. Citation2010), for example, suggests that without extensive pre- (and post-) release surveys it can be difficult to disentangle the effect of a transgenic trait, or the deliberate introduction of a new organism, from other variables that influence the desired outcome(s).
Variables that simultaneously influence: (a) which sites or potential sample units are exposed to a GDMO ‘treatment’, as the GDMO spreads through a population and (b) the outcome following a ‘treatment’, must be accounted for either through the monitoring design or in the analysis of the monitoring data or both, otherwise estimates of a treatment effect will be biased and interpretation of the causality of the effect will likely be flawed (Ferraro and Hanauer Citation2014; Dawid Citation2015).
A variety of design and analysis methods have been developed in an attempt to either eliminate or adjust for the effects of confounding variables in situations where randomized controlled trials are not possible (). Most of these methods were developed within medical domains, and some have been applied within environmental domains (Ferraro and Pressey Citation2015). All of these methods require: (i) at least a conceptual model of how the outcome of interest is likely to be influenced by observable and potentially unobservable variables; (ii) identification of an appropriate sampling frame and survey design and (iii) additional criteria or assumptions in order to assign a causal interpretation.
Table 3. Summary of methods to control for the effect of confounding variables, and help establish causality, with observational data.
An appropriate sampling frame and efficient survey design will be essential in any attempt to detect adverse outcomes and attribute these to a GDMO in Phase 4. The sample frame must allow for the spread and (at least temporary) persistence of the GDMO, and ideally extend to areas or regions where the degree of exposure to the GDMO will exhibit spatio-temporal gradients. Moreover, it is reasonable to anticipate that the analyst will know of some factors that will influence exposure to the GDMO, prior to its release, and if so the sampling frame should try to include a reasonable spread of the levels of these factors. For example, if the socio-economic status of neighborhoods or villages influences exposure to the GDMO, then it is important that the monitoring programme’s sampling frame does not exclude neighborhoods or villages from either end of the socio-economic spectrum.
Once the sample frame is established, randomized monitoring designs will help ensure that the sample data enables unbiased estimation of population parameters. Simple random designs and stratified random designs are traditionally used when monitoring ecological outcomes (McDonald Citation2010), such as those associated with open field release of GM fish (Senanan et al. Citation2007), but these types of design can often be improved through the use of spatially balanced designs (Stevens and Olsen Citation2004; Grafström and Lundström Citation2013). Spatially balanced designs are generally more efficient (provide more information per sample) than the traditional alternatives because they minimize spatial autocorrelation between samples.
The concept and advantages of spatial balance can also be extended into known or suspected covariates (Robertson et al. Citation2013). If covariates are included in a monitoring design, however, they should also be considered in the analysis (e.g. Gelman et al. Citation2013), and designs should only include covariates that are already known to strongly affect the observations. Including covariates of dubious explanatory power in the design will make the resulting model larger, and could reduce its ability to detect change. In the extreme case, if there are more covariates than observations in the classical linear model, the model will not have a unique set of estimates and inference becomes impossible.
In phase 4, the spatial and temporal scope of the analysis expands to community and ecosystem level scales. Prior to this stage, it will be useful to compare the efficiency of proposed monitoring designs under a variety of different assumptions regarding the spatio-temporal processes that govern factors such as dispersal and reproduction. For example, comparing a relatively simple process of gradual spread from a source that is slowly growing in size (Shigesda and Kawasaki Citation1997) against processes that include occasional long-range jumps (Marvier, Meir, and Kareiva Citation1999; Hallatscheka and Fisher Citation2014).
Additional considerations
The WHO phased testing and release pathway () can usefully support GDMO risk assessment. We would incorporate two modifications, however. The first is to note the importance of feedback between the Phases. Information and learning gathered during each of the phases should inform the design of experiments and analysis conducted in subsequent iterations of the pathway (NRC Citation1996). The second modification would be to add two additional Phases to address ‘entry’ and ‘exit’ strategies at the start and end of the pathway.
Entry strategies are deliberate activities that are conducted, prior to, or at least parallel with, Phase 1, and before a formal risk analysis process is initiated (NASEM Citation2016a calls these ‘Phase 0’ activities). These activities include enabling developers of gene drive technologies to work with interested and affected parties to develop a shared understanding of the problem that the GDMO could solve, setting standards or acceptability criteria for the performance of the GDMO in this context (e.g. through a Target Product Profile) and then comparing the GDMO solution with other options for solving the problem (Lynam et al. Citation2007; Nelson, Andow, and Banker Citation2009).
Exit strategies should define conditions under which the apparent absence of adverse ecological outcomes following an open field release is considered sufficient to cease monitoring and reporting. Interested and affected parties should also be involved in determining appropriate triggers for discontinuing monitoring programmes after open field release because this decision may be informed by a mix of social, political, or economic considerations as well as scientific evidence.
Entry strategies developed through a participatory process are expected to support fair and competent decisions to be made throughout the phased testing and release pathway: fair, in that the process helps to provide a role for interested or affected parties, and competent in that the process can facilitate the best decision possible given what was reasonably known under present conditions (Webler and Tuler Citation2000).
Hazard analysis techniques with a strong graphical basis, such as Qualitative Mathematical Modelling and Fault Tree Analysis (), are good ways to include a diverse set of interested and affected parties in hazard identification, including those with practical experience with relevant environmental systems. Here we emphasize the need for active participation of interested and affected parties throughout the analysis process, rather than simply communicating the results of the process to these parties. Efficacious stakeholder participation in the phased testing and release process will require a scientifically literate, neutral, and seasoned facilitator (Kaner et al. Citation2007).
Discussion
Risk analysis recommendations for the current generation of transgenic plants and fish emphasize the importance of adopting a case-by-case approach (NRC Citation2000; Rissler and Mellon Citation2000; Snow et al. Citation2005; Kapuscinski et al. Citation2007). We expect GDMOs to go through a similar case-by-case risk analysis process, informed by the type of gene-drive, the desired genotypic and phenotypic changes, and the characteristics of the environment into which the GDMO is being tested and released (NASEM Citation2016a).
The literature to date suggests that the potential adverse ecological outcomes following the deliberate or accidental release of GDMOs are the same as, or at least similar to, those identified for transgenic crops and fish, invasive species and biocontrol agents. The causal pathways (or chain of events) that lead to these potential outcomes, however, may be different because GDMOs contain genetic elements that are designed to spread a transgene across populations or subpopulations. We believe that structured hazard analysis tools will help researchers and risk analysts to systematically and comprehensively identify the causal pathways that lead to adverse environmental outcomes at population, community and ecosystem scales. The period immediately after the proof of concept of a transgenic construct (between phase 1 and 2 in ) is the appropriate time to conduct these hazard analysis studies and design experiments to detect, and screen for, intermediate events along the causal pathway.
If a decision is taken to move forward with the GDMO, then the process of identifying potential risks of the GDMO, risk management strategies for potential containment failures, and acceptance criteria for progression through each phase of the testing and release pathway should continue to involve relevant stakeholders (Dana et al. Citation2014). These considerations need to carefully describe the ecological or experimental outcomes that define a ‘successful’ GDMO at each Phase, the conditions under which adverse ecological outcomes would be considered significant enough to terminate development of the GDMO, and the conditions under which the apparent absence of adverse ecological outcomes is sufficient to terminate post-release monitoring.
Long-term monitoring strategies for unexpected adverse ecological outcomes may also (depending on the GDMO application) require innovations in monitoring platforms, such as cheap, automated trapping, and governance and funding arrangements that are secure enough to withstand changing policy and budget priorities. Furthermore, the detection of adverse ecological outcomes may need to be accompanied by additional analysis or experiments to determine the mechanisms by which such an event could be attributable to the GDMO.
Whilst this paper focuses on identifying hazards and detecting potential adverse ecological outcomes associated with GDMOs, it is important to not lose sight of the benefits that gene drive technologies may offer (NASEM Citation2016a). We recognize that any decision to deliberately test and release GDMOs must balance the benefits and risks of the technology, and compare these with alternative solutions (Nelson, Andow, and Banker Citation2009). The key issue now is how to integrate transparently developed, scientifically testable, hazard identification and risk assessment methods, within a fair and inclusive decision-making framework that clearly enunciates the risk-based acceptance criteria for progressing this new technology through a phased test and release pathway.
Acknowledgements
The authors would like to thank two anonymous reviewers for their thoughtful and helpful comments, as well as the organizers of the NCSU gene drive workshop for the opportunity to contribute to the ‘Road Map to Gene Drive’ special issue.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes on contributors
Keith R. Hayes is a Senior Research Scientist in CSIRO. Keith leads a small team of statisticians and modellers that focus on developing and applying probabilistic risk assessment methods in the ecological domain.
Geoffrey R. Hosack is a Research Scientist at CSIRO working with the CSIRO’s Environment and Ecological Risk team. Geoff’s research focuses on integrating data analytic methods, monitoring techniques and expert elicitation within risk analyses applied to the domains of ecosystem modelling, genetic engineering and vector-borne disease control.
Genya V. Dana is trained in risk analysis and works on international science and technology policy issues, with a focus on emerging biotechnologies.
Scott D. Foster is a Senior Research Scientist with the CSIRO in Hobart, Australia. The applications are what interests Scott, as well as the (sometimes novel) statistical methods to address them. The applications he works on are quite diverse and include: biodiversity quantification, community modelling, environmental modelling, genetics and fisheries science.
Jessica H. Ford is a Research Scientist working with the CSIRO’s Environment and Ecological Risk team. She is an applied statistician working in areas of environmental risk, fisheries management and the genetic control of vector-borne diseases.
Ron Thresher was the foundation head of the CSIRO Centre for Research on Introduced Marine Pests, and has lead genetic programmes to limit the ecological impacts of escapees from aquaculture facilities (‘sterile feral’ technology) and to manage lower vertebrate pests by means of ‘daughterless’ heritable sex-ratio biasing.
Adrien Ickowicz is a Research Statistician working with the CSIRO’s Environment and Ecological Risk team. His research interest includes spatial–temporal modeling and monitoring, computational and Bayesian statistics.
David Peel is a research scientist with the CSIRO in Hobart, Australia. He is a statistician with a strong application focus specializing in ecological and environmental areas.
Mark Tizard is a team leader at CSIRO Health and Biosecurity. His primary research interest is in genetic control of vertebrate pests.
Paul De Barro is Senior Principal Research Scientist and Research Director of the Risk Evaluation and Preparedness Program at CSIRO Health and Biosecurity.
Tanja Strive is a Principal Research Scientist at CSIRO Health and Biosecurity. Her research interests are in biological control of vertebrates and host–pathogen interactions and co-evolution. Previous research experience ranging from non-lethal, virally vectored immune-contraception approaches for foxes and mice to classical viral biocontrol of rabbits using naturally occurring pathogens.
Jeffrey M. Dambacher is a Senior Research Scientist working with the CSIRO’s Environment and Ecological Risk team. He specializes in the application of Qualitative Mathematical Modelling to problems in the ecological domain.
ORCID
Jessica H. Ford http://orcid.org/0000-0003-4473-8245
Adrien Ickowicz http://orcid.org/0000-0001-7861-7340
Mark Tizard http://orcid.org/0000-0001-8421-2497
References
- ACME/ASTMH (American Committee of Medical Entomology and the American Society of Tropical Medicine and Hygiene). 2003. “Arthropod Containment Guidelines. A Project of the American Committee of Medical Entomology and American Society of Tropical Medicine and Hygiene.” Vector Borne and Zoonotic Diseases 3: 61–98. doi: 10.1089/153036603322163448
- Akbari, O. S., H. J. Bellen, E. Bier, S. L. Bullock, A. Burt, G. M. Church, K. R. Cook, et al. 2015. “Safeguarding Gene Drive Experiments in the Laboratory.” Science 349: 927–929. doi:10.1126/science.aac7932.
- Andow, D. A., and C. Zwahlen. 2006. “Assessing Environmental Risks of Transgenic Plants.” Ecology Letters 9: 196–214. doi: 10.1111/j.1461-0248.2005.00846.x
- Ankley, G. T., R. S. Bennett, R. J. Erickson, D. J. Hoff, M. W. Hornung, R. D. Johnson, D. R. Mount, et al. 2010. “Adverse Outcome Pathways: a Conceptual Framework to Support Ecotoxicology Research and Risk Assessment.” Environmental Toxicology and Chemistry 29: 730–741. doi:10.1002/etc.34.
- Araki, M., K. Nojima, and T. Ishii. 2014. “Caution Required for Handling Genome Editing Technology.” Trends in Biotechnology 32: 234–237. doi: 10.1016/j.tibtech.2014.03.005
- Aven, T., and T. E. Nøkland. 2010. “On the use of Uncertainty Importance Measures in Reliability and Risk Analysis.” Reliability Engineering and System Safety 95: 127–133. doi: 10.1016/j.ress.2009.09.002
- Barratt, B. I. P., F. G. Howarth, T. M. Withers, J. M. Kean, and G. S. Ridley. 2010. “Progress in Risk Assessment for Classical Biological Control.” Biological Control 52: 245–254. doi:10.1016/j.biocontrol.2009.02.012.
- Benedict, M., A. Burt, M. L. Capurro, P. De Barro, A. M. Handler, K. R. Hayes, J. M. Marshall, W. J. Tabachnick, and Z. N. Adelman. 2017. “Management and Containment in Laboratories Handling Invasive Genetic Factors in Arthropods.” Vector Borne and Zoonotic Diseases. doi:10.1089/vbz.2017.2121.
- Botstein, D., S. A. Chervitz, and M. Cherry. 1997. “Yeast as a Model Organism.” Science 277: 1259–1260. doi:10.1126/science.277.5330.1259.
- Braig, H. R., and G. Yan. 2002. “The Spread of Genetic Constructs in Natural Insect Populations.” In Genetically Engineered Organisms: Assessing Environmental and Human Health Effects, edited by D. K. Letourneau, and B. E. Burrows, 251–314. Boca Raton, FL: CRC Press.
- Burt, A. 2003. “Site-specific Selfish Genes as Tools for the Control and Genetic Engineering of Natural Populations.” Proceedings of the Royal Society B: Biological Sciences 270: 921–928. doi:10.1098/rspb.2002.2319.
- Burt, A. 2014. “Heritable Strategies for Controlling Insect Vectors of Disease.” Philosophical Transactions of the Royal Society B: Biological Sciences 369: 20130432. doi: 10.1098/rstb.2013.0432
- Burt, A., M. Coulibaly, A. Crisanti, A. Diabate, and J. K. Kayondo. 2018. “ Gene Drive to Reduce Malaria Transmission in Sub-Saharan Africa”. Journal of Responsible Innovation 5 (S1): S66–S80.
- Champer, J., A. Buchman, and O. S. Akbari. 2016. “Cheating Evolution: Engineering Gene Drives to Manipulate the Fate of Wild Populations.” Nature Reviews Genetics 17 (3): 146–159. doi: 10.1038/nrg.2015.34
- Dambacher, J. M., H. W. Li, and P. A. Rossignol. 2003. “Qualitative Predictions in Model Ecosystems.” Ecological Modelling 161: 79–93. doi: 10.1016/S0304-3800(02)00295-8
- Dana, G. V., A. M. Cooper, K. M. Pennington, and L. S. Sharpe. 2014. “Methodologies and Special Considerations for Environmental Risk Analysis of Genetically Modified Aquatic Biocontrol Organisms.” Biological Invasions 16: 1257–1272. doi:10.1007/s10530-012-0391-x.
- David, A. S., J. M. Kaser, A. C. Morey, A. M. Roth, and D. A. Andow. 2013. “Release of Genetically Engineered Insects: A Framework to Identify Potential Ecological Effects.” Ecology and Evolution 3 (11): 4000–4015. doi: 10.1002/ece3.737
- Dawid, A. P. 2015. “Statistical Causality from a Decision-Theoretic Perspective.” Annual Review of Statistics and Its Application 2: 273–303. doi: 10.1146/annurev-statistics-010814-020105
- Devlin, R. H., L. F. Sundstrom, J. I. Johnsson, I. A. Fleming, K. R. Hayes, W. O. Ojwang, C. Bambaradeniya, and M. Zakaraia-Ismail. 2007. “Assessing Ecological Effects of Transgenic Fish Prior to Entry into Nature.” In Environmental Risk Assessment of Genetically Modified Organisms. Volume 3: Methodologies for Transgenic Fish, edited by A. R. Kapuscinski, K. R. Hayes, S. Li, and G. Dana, 151–187. Oxfordshire: CABI.
- DiCarlo, J. E., A. Chavez, S. L. Dietz, K. M. Esvelt, and G. M. Church. 2015. “Safeguarding CRISPR-Cas9 Gene Drives in Yeast.” Nature Biotechnology 33: 1250–1255. doi: 10.1038/nbt.3412
- Doudna, J. 2015. “Genome-editing Revolution: My Whirlwind Year with CRISPR.” Nature 528: 469–471. doi: 10.1038/528469a
- Duan, J. J., J. G. Lundgren, S. Naranjo, and M. Marvier. 2009. “Extrapolating Non-target Risk of Bt Crops from Laboratory to Field.” Biology Letters. doi:10.1098/rsbl.2009.0612.
- Ericson, C. A. 2005. Hazard Analysis Techniques for System Safety. Hoboken, NJ: John Wiley.
- Esvelt, K. M., A. L. Smidler, F. Catteruccia, and G. M. Church. 2014. “Concerning RNA-guided Gene Drives for the Alteration of Wild Populations.” eLife 3. doi:10.7554/eLife.03401.
- Facchinelli, L., L. Valerio, J. M. Ramsey, F. Gould, R. K. Walsh, G. Bond, M. A. Robert, et al. 2013. “Field Cage Studies and Progressive Evaluation of Genetically-Engineered Mosquitoes.” PLOS Neglected Tropical Diseases 7: e2001. doi:10.1371/journal.pntd.0002001.
- Ferguson, H. M., K. R. Ng’habi, T. Walder, D. Kadungula, S. J. Moore, I. Lyimo, T. L. Russell, et al. 2008. “Establishment of a Large Semi-Field System for Experimental Study of African Malaria Vector Ecology and Control in Tanzania.” Malaria Journal 158: 1475–2875.
- Ferraro, P. J., and M. M. Hanauer. 2014. “Advances in Measuring the Environmental and Social Impacts of Environmental Programs.” Annual Review of Environment and Resources 39: 495–517. doi:10.1146/annurevenviron-101813-013230 doi: 10.1146/annurev-environ-101813-013230
- Ferraro, P. J., and R. L. Pressey. 2015. “Measuring the Difference Made by Conservation Initiatives: Protected Areas and Their Environmental and Social Impacts.” Philosophical Transactions of the Royal Society, Series B 370. doi:10.1098/rstb.2014.0270.
- Firbank, L., M. Lonsdale, and G. Poppy. 2005. “Re-assessing the Environmental Risks of GM Crops.” Nature Biotechnology 23: 1475–1476. doi: 10.1038/nbt1205-1475
- Ford, J. H., K. R. Hayes, B. L. Henderson, S. Lewis, and P. Baker. 2015. Systematic Analysis of Water-Related Hazards Associated with Coal Resource Development. Sub-Methodology M11 from the Bioregional Assessment Technical Programme. Canberra: Department of the Environment, Bureau of Meteorology, CSIRO and Geoscience Australia.
- Gaj, T., C. A. Gersbach, and C. F. Barbas. 2014. “ZFN, TALEN, and CRISPR/Cas-based Methods for Genome Engineering.” Trends in Biotechnology 31: 397–405. doi:10.1016/j.tibtech.2013.04.004.
- Gantz, V. M., and E. Bier. 2015. “The Mutagenic Chain Reaction: A Method for Converting Heterozygous to Homozygous Mutations.” Science 348: 442–444. doi:10.1126/science.aaa5945.
- Gantz, V. M., N. Jasinskiene, O. Tatarenkova, A. Fazekas, V. M. Macias, E. Bier, and A. A. James. 2015. “Highly Efficient Cas9-Mediated Gene Drive for Population Modification of the Malaria Vector Mosquito Anopheles Stephensi.” Proceedings of the National Academy of Sciences 112: E6736–E6743. doi:10.1073/pnas.1521077112.
- Gelman, A., J. B. Carlin, H. S. Stern, D. B. Dunson, A. Vehtari, and D. B. Rubin. 2013. Bayesian Data Analysis. 3rd ed.Boca Raton, FL: Chapman and Hall.
- Gerlach, R., K. Mengersen, and F. Tuyl. 2009. “Posterior Predictive Arguments in Favor of the Bayes-Laplace Prior as the Consensus Prior for Binomial and Multinomial Parameters.” Bayesian Analysis 4: 151–158. doi: 10.1214/09-BA405
- Gould, F., K. Magori, and Y. Huang. 2006. “Genetic Strategies for Controlling Mosquito-Borne Diseases.” American Scientist 94: 238–246. doi:10.1511/2006.3.238 doi: 10.1511/2006.59.992
- Grafström, A., and N. L. P. Lundström. 2013. “Why Well Spread Probability Samples Are Balanced.” Open Journal of Statistics 3: 36–41. doi:10.4236/ojs.2013.31005.
- Grimes, D. A., and K. F. Schulz. 2002. “Bias and Causal Associations in Observational Research.” The Lancet 359: 248–252. doi: 10.1016/S0140-6736(02)07451-2
- Hallatscheka, O., and D. S. Fisher. 2014. “Acceleration of Evolutionary Spread by Long-Range Dispersal.” Proceedings of the National Academy of Sciences 111: E4911–E4919. doi:10.1073/pnas.1404663111.
- Hammond, A., R. Galizi, K. Kyrou, A. Simoni, C. Siniscalchi, D. Katsanos, M. Gribble, et al. 2015. “A CRISPR/Cas9 Gene Drive System Targeting Female Reproduction in the Malaria Mosquito Vector Anopheles gambiae.” Nature Biotechnology 34: 78–83. doi:10.1038/nbt.3439.
- Hawes, C., A. J. Haughton, J. L. Osborne, D. B. Roy, S. J. Clark, J. N. Perry, P. Rothery, et al. 2003. “Responses of Plants and Invertebrate Trophic Groups to Contrasting Herbicide Regimes in the Farm Scale Evaluations of Genetically Modified Herbicide-Tolerant Crops.” Philosophical Transactions of the Royal Society B: Biological Sciences 358: 1899–1913. doi:10.1098/rstb.2003.1406.
- Hayes, K. R. 2002a. “Identifying Hazards in complex Ecological Systems, Part 1: Fault Tree Analysis for Biological Invasions.” Biological Invasions 4 (3): 235–249. doi: 10.1023/A:1020979914453
- Hayes, K. R. 2002b. “Identifying Hazards in complex Ecological Systems, Part 2: Infections Modes and Effects Analysis for Biological Invasions.” Biological Invasions 4 (3): 251–261. doi: 10.1023/A:1020943231291
- Hayes, K. R., S. C. Barry, N. Beebe, J. M. Dambacher, P. De Barro, S. Ferson, J. Ford, et al. 2015. Risk Assessment for Controlling Mosquito Vectors with Engineered Nucleases, Sterile Male Construct: Final Report. CSIRO Biosecurity Flagship, Hobart, Australia, 202. Accessed March 21, 2016. http://targetmalaria.org/resources/.
- Hayes, K. R., P. C. Gregg, V. V. S. R. Gupta, R. Jessop, M. Lonsdale, B. Sindel, J. Stanley, and C. K. Williams. 2004. “Identifying Hazards in complex Ecological Systems. Part 3: Hierarchical Holographic Model for Herbicide Tolerant Oilseed Rape.” Environmental BioSafety Research 3: 1–20. doi: 10.1051/ebr:2004012
- Hayes, K. R., B. Leung, R. E. Thresher, J. Dambacher, and G. Hosack. 2014. “Assessing the Risks of Genetic Control Techniques with Reference to the Common Carp (Cyprinus carpio) in Australia.” Biological Invasions 16: 1273–1288. doi:10.1007/s10530-012-0392-9.
- Hilbeck, A., D. Andow, E. M. G. Fontes, A. R. Kapuscinski, and P. J. Schei, eds. 2005. Environmental Risk Assessment of Genetically Modified Organisms, Volume 2: Methodologies for Assessing Bt Cotton in Brazil. Wallingford: CABI. 373.
- Hunt, G. J., and W. J. Tabachnick. 1996. “Handling Small Arbovirus Vectors Safely During Biosafety Level 3 Containment: Culicoides variipennis sonorensis (Diptera: Ceratopogonidae) and Exotic Bluetongue Viruses.” Journal of Medical Entomology 33: 271–277. doi: 10.1093/jmedent/33.3.271
- Iman, R. L. 1987. “A Matrix-Based Approach to Uncertainty and Sensitivity Analysis for Fault Trees.” Risk Analysis 7: 21–33. doi: 10.1111/j.1539-6924.1987.tb00966.x
- Jinek, M., K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna, and E. Charpentier. 2012. “A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity.” Science 337: 816–821. doi:10.1126/science.1225829.
- Kaner, S., L. Lind, C. Toldi, S. Fisk, and D. Berger. 2007. Facilitator’s Guide to Participatory Decision-making. San Francisco, CA: Jossey-Bass.
- Kapuscinski, A. R., G. Dana, K. R. Hayes, S. Li, K. C. Nelson, Y. K. Nam, Z. Gong, R. H. Devlin, G. C. Mair, and W. Senanan. 2007. “Risk Assessment of Transgenic Fish: Synthesis and Conclusions.” In Environmental Risk Assessment of Genetically Modified Organisms. Volume 3: Methodologies for Transgenic Fish, edited by A. R. Kapuscinski, K. R. Hayes, S. Li, and G. Dana, 272–289. Oxfordshire: CABI.
- Knols, B., B. Njiru, R. Mukabana, E. Methenge, and G. Killeen. 2003. “Contained Semi-Field Environments for Ecological Studies on Transgenic African Malaria Vectors: Benefits and Constraints.” In Ecological Aspects for Application of Genetically Modified Mosquitoes, edited by W. Takken, and T. W. Scott, 91–106. New York: Springer-Verlag.
- Kumamoto, H., and E. Henley. 1996. Probabilistic Risk Assessment and Management for Engineers and Scientists. Piscataway, NJ: IEEE Press. 597.
- Le Cong, F., A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, et al. 2013. “Multiplex Genome Engineering Using CRISPR/Cas Systems.” Science 339: 819–823. doi:10.1126/science.1231143.
- Leitschuh, C., D. Kanavy, G. Backus, D. Threadgill, and J. Godwin. 2017. “ Eradicating invasive rodents from islands: An assessment of current and future gene drive technologies”. Journal of Responsible Innovation 5 (S1): S121–S138.
- Lindholm, A. K., K. A. Dyer, R. C. Firman, L. Fishman, W. Forstmeier, L. Holman, H. Johannesson, et al. 2016. “The Ecology and Evolutionary Dynamics of Meiotic Drive.” Trends in Ecology and Evolution 31: 315–326. doi:10.1016/j.tree.2016.02.001.
- Lobo, I. 2008. “Environmental Influences on Gene Expression.” Nature Education 1 (1): 39.
- Lövei, G. L., and S. Arpaia. 2004. “The Impact of Transgenic Plants on Natural Enemies: A Critical Review of Laboratory Studies.” Entomologia Experimentalis et Applicata 114: 1–14. doi:10.1111/j.0013-8703.2005.00235.x.
- Lynam, T., W. de Jong, D. Sheil, T. Kusumanto, and K. Evans. 2007. “A Review of Tools for Incorporating Community Knowledge, Preferences, and Values into Decision Making in Natural Resources Management.” Ecology and Society 12: 5. http://www.ecologyandsociety.org/vol12/iss1/art5/. doi: 10.5751/ES-01987-120105
- Madigan, D., P. E. Stang, J. A. Berlin, M. Schuemie, J. M. Overhage, M. A. Suchard, B. Dumouchel, A. G. Hartzema, and P. B. Ryan. 2014. “A Systematic Statistical Approach to Evaluating Evidence from Observational Studies.” Annual Review of Statistics and Its Application 1: 11–39. doi:10.1146/annurev-statistics-022513-115645.
- Marvier, M. A., E. Meir, and P. M. Kareiva. 1999. “How do the Design of Monitoring and Control Strategies Affect the Chance of Detecting and Containing Transgenic Weeds?” In Methods for Risk Assessment of Transgenic Plants III. Ecological Risks and Prospects of Transgenic Plants, edited by K. Ammann, Y. Jacot, V. Simonsen, and G. Kjellsson, 109–122. Basel: Birkhauser Verlag.
- McDonald, T. 2010. “Spatial Sampling Designs for Long-Term Ecological Monitoring.” In Design and Analysis of Long-term Ecological Monitoring Studies, edited by R. A. Gitzen, J. J. Millspaugh, A. B. Cooper, and D. S. Licht, 101–125. Cambridge: Cambridge University Press.
- Moe-Behrens, G. H. G., R. Davis, and K. A. Haynes. 2013. “Preparing Synthetic Biology for the World.” Frontiers in Microbiology 4: 1–10. doi:10.3389/fmicb.2013.00005.
- Muir, W. M., and R. D. Howard. 2002. “Assessment of Possible Ecological Risks and Hazards of Transgenic Fish with Implications for Other Sexually Reproducing Organisms.” Transgenic Research 11: 101–114. doi: 10.1023/A:1015203812200
- Mutze, G., B. Cooke, and P. Alexander. 1995. “The Initial Impact of Rabbit Hemorrhagic Disease on European Rabbit Populations in South Australia.” Journal of Wildlife Diseases 34: 221–227. doi:10.7589/0090-3558-34.2.221.
- National Academy of Sciences Engineering and Medicine. 2016a. Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty and Aligning Research with Public Values. Washington, DC: The National Academies Press. doi:10.17226/23405.
- National Academy of Sciences Engineering and Medicine. 2016b. Genetically Engineered Crops: Experiences and Prospects. Washington, DC: The National Academies Press. doi:10.17226/23395.
- Nelson, K. C., D. A. Andow, and M. J. Banker. 2009. “Problem Formulation and Option Assessment (PFOA) Linking Governance and Environmental Risk Assessment for Technologies: A Methodology for Problem Analysis of Nanotechnologies and Genetically Engineered Organisms.” Journal of Law, Medicine and Ethics 37: 732–748. doi:10.1111/j.1748-720X.2009.00444.x.
- Nguyen, T. H., H. L. Nguyen, T. Y. Nguyen, S. N. Vu, N. D. Tran, T. N. Le, Q. M. Vien, et al. 2015. “Field Evaluation of the Establishment Potential of Wmelpop Wolbachia in Australia and Vietnam for Dengue Control.” Parasites and Vectors 8: 1756–3305. doi:10.1186/s13071-015-1174-x.
- NRC (National Research Council). 1996. Understanding Risk: Informing Decisions in a Democratic Society. Washington, DC: National Academies Press. 264.
- NRC (National Research Council). 2000. Genetically Modified Pest-Protected Plants; Science and Regulation. Washington, DC: National Academies Press. 263.
- Pennington, K. M., A. R. Kapuscinski, M. S. Morton, A. M. Copper, and L. M. Miller. 2010. “Full Life-Cycle Assessment of Gene Flow Consistent with Fitness Differences in Transgenic and Wild-type Japanese Medaka Fish (Oryzias latipes).” Environmental Biosafety Research 9: 41–57. doi:10.1051/ebr/2010005.
- Rausand, M., and A. Hoyland. 2004. System Reliability Theory: Models, Statistical Methods and Applications. 2nd ed.London: Wiley.
- Renn, O. 2008. Risk Governance: Coping with Uncertainty in a Complex World. London: Routledge.
- Rissler, J., and M. Mellon. 2000. The Ecological Risks of Engineered Crops. Cambridge, MA: The MIT Press. 168.
- Robertson, B. L., J. A. Brown, T. McDonald, and P. Jaksons. 2013. “BAS: Balanced Acceptance Sampling of Natural Resources.” Biometrics 69: 776–784. doi:10.1111/biom.12059.
- Royal Society. 1983. Risk Assessment. Report of a Royal Society Study Group. London: The Royal Society. 75.
- Rubin, D. B. 1974. “Estimating Causal Effects of Treatments in Randomized and Nonrandomized Studies.” Journal of Educational Psychology 66: 688–701. doi: 10.1037/h0037350
- Saey, T. H. 2015. “Gene Drives Spread Their Wings.” Science News 188: 16.
- Sander, J. D., and J. K. Joung. 2014. “CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes.” Nature Biotechnology 32: 347–355. doi:10.1038/nbt.2842.
- Scott, M., F. Gould, M. Lorenzen, N. Grubbs, O. Edwards, and D. O’Brochta. 2017. “ Agricultural Production: Assessment of the Potential use of Cas9-mediated Gene Drive Systems for Agricultural Pest Control”. Journal of Responsible Innovation 5 (S1): S98–S120.
- Scott, T. W., W. Takken, B. G. J. Knols, and C. Boete. 2002. “The Ecology of Genetically Modified Mosquitoes.” Science 298: 117–119. doi: 10.1126/science.298.5591.117
- Senanan, W., J. J. Hard, A. Alcivar-Warren, J. Trisak, M. Zakaraia-Ismail, and M. L. Hernandez. 2007. “Risk Management: Post-Approval Monitoring and Remediation.” In Environmental Risk Assessment of Genetically Modified Organisms. Volume 3: Methodologies for Transgenic Fish, edited by A. R. Kapuscinski, K. R. Hayes, S. Li, and G. Dana, 239–271. Oxfordshire: CABI.
- Shigesda, N., and K. Kawasaki. 1997. Biological Invasions: Theory and Practice. Oxford: Oxford University Press.
- Simberloff, D. 2012. “Risks of Biological Control for Conservation Purposes.” Biocontrol 57: 263–276. doi: 10.1007/s10526-011-9392-4
- Simberloff, D., J. L. Martin, P. Genovesi, V. Maris, D. A. Wardle, J. Aronson, F. Courchamp, et al. 2013. “Impacts of Biological Invasions: What’s What and the Way Forward.” Trends in Ecology and Evolution 28: 58–66. doi: 10.1016/j.tree.2012.07.013
- Smidler, A., J. Min, and K. Esvelt. 2017. “ Harnessing gene drive systems”. Journal of Responsible Innovation 5 (S1): S40–S65.
- Snow, A. A., D. A. Andow, P. Gepts, E. M. Hallerman, A. Power, J. M. Tiedje, and L. L. Wolfenbarger. 2005. “Genetically Engineered Organisms and the Environment: Current status and Recommendations.” Ecological Applications 15: 377–404. doi: 10.1890/04-0539
- Stevens, D. L., and A. R. Olsen. 2004. “Spatially Balanced Sampling of Natural Resources.” Journal of the American Statistical Association 99: 262–278. doi:10.1198/016214504000000250.
- Sundström, L. F., M. Lõhmus, W. E. Tymchuk, and R. H. Devlin. 2007. “Gene–Environment Interactions Influence Ecological Consequences of Transgenic Animals.” Proceedings of the National Academy of Sciences 104: 3889–3894. doi: 10.1073/pnas.0608767104
- Tabachnick, W. J. 2003. “Reflections on the Annopheles gambiae Genome Sequence, Transgenic Mosquitoes and the Prospect for Controlling Malaria and Other Vector Borne Diseases.” Journal of Medical Entomology 40: 597–606. doi:10.1603/0022-2585-40.5.597.
- Tsai, S. Q., Z. Zheng, N. T. Nguyen, M. Liebers, V. V. Topkar, V. Thapar, N. Wyvekens, et al. 2015. “GUIDE-seq Enables Genome-Wide Profiling of off-Target Cleavage by CRISPR-Cas Nucleases.” Nature Biotechnology 33: 187–197. doi:10.1038/nbt.3117.
- Watts, S. 1989. “Britons Pioneer Assessment of Gene Hazards.” New Scientist 1666: 32.
- Webb, J. L. A. 2011. “The First Large-Scale Use of Synthetic Insecticide for Malaria Control in Tropical Africa: Lessons from Liberia, 1945–1962.” Journal of the History of Medicine and Allied Sciences 66: 347–376. doi: 10.1093/jhmas/jrq046
- Webber, B. L., S. Raghu, and O. R. Edwards. 2015. “Is CRISPR-based Gene Drive a Biocontrol Silver Bullet or Global Conservation Threat?” Proceedings of the National Academy of Sciences 112: 10565–10567. doi:10.1073/pnas.1514258112.
- Webler, T., and S. Tuler. 2000. “Fairness and Competence in Citizen Participation.” Administration and Society 32: 566–595. doi: 10.1177/00953990022019588
- WHO (World Health Organisation). 2014. Guidance Framework for Testing Genetically Modified Mosquitoes. Technical Report, World Health Organisation, Special Programme for Research and Training in Tropical Diseases. Accessed November 23, 2017. http://www.who.int/tdr/publications/year/2014/guide-fmrk-gm-mosquit/en/.
- WHO (World Health Organization). 2009. Progress and Prospects for the Use of Genetically Modified Mosquitoes to Inhibit Disease Transmission. Technical Report, World Health Organisation, Special Programme for Research and Training in Tropical Diseases. Accessed November 23, 2017. http://www.who.int/tdr/publications/training-guideline-publications/gmm-report/en/.
- Wijayawardena, B. K., D. J. Minchella, and J. A. DeWoody. 2013. “Hosts, Parasites, and Horizontal Gene Transfer.” Trends in Parasitology 29: 329–338. doi: 10.1016/j.pt.2013.05.001
- Wilson, E. B. 1927. “Probable Inference, the law of Succession, and Statistical Inference.” Journal of the American Statistical Association 22: 209–212. doi: 10.1080/01621459.1927.10502953
- Winkler, R. L., J. E. Smith, and D. G. Fryback. 2002. “The Role of Informative Priors in Zero-Numerator Problems.” The American Statistician 56: 1–4. doi: 10.1198/000313002753631295
- Wood, R. J., and M. E. Newton. 1991. “Sex-ratio Distortion Caused by Meiotic Drive in Mosquitoes.” The American Naturalist 137: 379–391. doi: 10.1086/285171
- Wu, B., L. Luo, and X. J. Gao. 2016. “Cas9-triggered Chain Ablation of cas9 as a Gene Drive Brake.” Nature Biotechnology 34: 137–138. doi: 10.1038/nbt.3444
- Zimmering, S., L. Sandler, and B. Nicoletti. 1970. “Mechanisms of Meiotic Drive.” Annual Review of Genetics 4 (1): 409–436. doi: 10.1146/annurev.ge.04.120170.002205
