Abstract
Vitamin A deficiency is a global health problem and can be effectively alleviated through crop biofortification. Quantification of carotenoids using high-performance liquid chromatography is expensive and time-consuming, thereby posing a challenge in the selection of genotypes with high provitamin A. Favourable alleles possessing rare genetic variation in lycopene-ε-cyclase (lcyE) and β-carotene hydroxylase (crtRB1) genes are associated with higher accumulation of provitamin A, especially β-carotene; and selection of these alleles holds immense promise in reducing large-scale phenotypic assays. Screening of a diverse set of 385 maize inbred lines of indigenous and exotic origin detected the presence of two alleles (amplicon size: 250 and 650 bp) of lcyE and three alleles (amplicon size: 296, 543 and 875 bp) of crtRB1 in the inbred panel. Favourable alleles of both the genes were rare among the traditional maize germplasm; 3.38% of the inbreds possessed the favourable allele (650 bp) of lcyE, while 3.90% inbreds had the favourable allele (543 bp) of crtRB1. Five inbreds (1.3%) with favourable alleles of both the genes were found. Inbreds with favourable alleles of crtRB1 and lcyE serve as rich genetic resources for effective utilization in the maize biofortification programme.
Public Interest Statement
The work presented in the manuscript describes the promise of maize inbreds with favourable alleles of lycopene-ε-cyclase and β-carotene hydroxylase genes for enrichment of kernel β-carotene in maize. This also illustrates its utility for diversification of maize germplasm for kernel carotenoids. The inbreds with the favourable allele and high β-carotene serve as potential genetic resources that can be used for the accelerated development of maize hybrids with high β-carotene using marker-assisted selection. These biofortified maize genotypes will serve as sustainable and cost-effective solutions to alleviate micronutrient malnutrition worldwide.
1. Introduction
Micronutrient malnutrition or “hidden hunger” has become a major health problem and affects millions of people worldwide. Among them, vitamin A deficiency (VAD) alone affects over 250 million pre-school children and accounts for about 70% of the childhood deaths globally (Vignesh et al., Citation2012). Young children, pregnant women and lactating mothers are most vulnerable to VAD, as it contributes to maternal death, poor pregnancy and lactation (Black et al., Citation2008). Besides, it also contributes to the predisposition of several major diseases, such as anaemia, diarrhoea, measles, malaria and respiratory infections (West, Citation2000). Since vitamin A cannot be synthesized inside the human body, it needs to be provided through diet. Hence, increasing provitamin A carotenoids in staple crops through breeding would be a viable strategy to minimize the adverse effects of VAD (Bouis, Hotz, McClafferty, Meenakshi, & Pfeiffer, Citation2011).
As compared to other staple cereals, maize possesses tremendous variability for kernel carotenoids (Buckner, Kelson, & Robertson, Citation1990). Quantifying the provitamin A carotenoid using high-performance liquid chromatography (HPLC) is expensive and time-consuming. Polymerase chain reaction (PCR)-based assay thus would help in identifying genotypes with high β-carotene. Three genes in the carotenoid biosynthesis pathway play a crucial role in the accumulation of various carotenoids in the maize kernel. The first committed step in the pathway is regulated by phytoene synthase (psy) that converts phytoene to geranylgeranyl pyrophosphate leading to yellow/orange-coloured maize endosperm (Buckner et al., Citation1990). The first branching point of the pathway is the cyclization of lycopene: lycopene-ε-cyclase (lcyE) gene, in association with other genes, converts more lycopene to the β, ε branch, which produces more α-carotene and lutein (Harjes et al., Citation2008). Another key gene in the pathway is β-carotene hydroxylase (crtRB1) that causes the hydroxylation of α- and β-carotene into the non-provitamin A carotenoids viz. lutein and zeaxanthin, respectively (Vallabhaneni et al., Citation2009; Yan et al., Citation2010). Therefore, pathway branching and hydroxylation are key determinants in controlling provitamin A levels of the maize kernel (Harjes et al., Citation2008; Yan et al., Citation2010).
Harjes et al. (Citation2008) showed that naturally existing mutant allele at the lycE locus alters flux down α-carotene versus β-carotene branches of the carotenoid biosynthesis pathway. Using allele mining strategy, four natural lcyE polymorphisms viz. lcyE 5′TE (Transposable Element in 5′-untranslated region [UTR]), lcyE SNP216 (in exon 1), lcyE SNP2238 (in intron 4) and lcyE 3′InDel (in 3′-UTR) were identified, of which, the favourable allele of lcyE 5′TE causes more increase in provitamin A content (Harjes et al., Citation2008). Harjes et al. (Citation2008) reported the presence of four alleles in the 5′TE region of the lcyE gene viz. allele 1 (150 + 280 bp), allele 2 (250 bp), allele 3 (250 bp + 380 bp) and allele 4 (650 bp). Of these, allele 1 and allele 4 are known as favourable alleles for enhancing the β-branch carotenoids by increasing the pathway flux towards β- branch, whereas allele 2 and allele 3 cause unfavourable effects. Yan et al. (Citation2010) through association mapping approach detected three polymorphisms viz. 5′TE (in the 5′-UTR), InDel4 (in the coding region) and 3′TE (spanning the sixth exon and 3′-UTR) in crtRB1 that were significantly associated with the β-carotene concentration and its conversion to β-cryptoxanthin and zeaxanthin in maize kernels (Yan et al., Citation2010), of which crtRB1 3′TE favourable allele alone caused two to tenfold variations in the β-carotene concentration among the inbreds (Babu, Rojas, Gao, Yan, & Pixley, Citation2013; Muthusamy et al., Citation2014). The 3′TE polymorphism of crtRB1 gene that spans the 6th exon and the 3′-UTR that generates three alleles viz. allele 1 (543 bp; without TE insertion), allele 2 (296 bp + 875 bp; with 325 bp TE insertion) and allele 3 (296 bp + 1221 bp + 1880 bp; with 1250 bp TE insertion) were associated with altering β-carotene accumulation (Yan et al., Citation2010). Allele 1 is known as a favourable allele for enhancing the β-carotene by reducing transcript expression of the crtRB1 gene, whereas allele 2 and allele 3 cause unfavourable effects. PCR-based co-dominant markers were identified for both lcyE- and crtRB1-based polymorphisms which can pave way for rapid improvement of provitamin A content in maize kernels through marker-assisted selection (MAS) (Zhang et al., Citation2012). Thus, the present study was undertaken to screen a large set of inbreds of indigenous and exotic origin to identify genotypes having favourable alleles of lcyE and crtRB1, for their utilization in maize provitamin A biofortification programme.
2. Materials and methods
2.1. Plant material
A diverse set of 385 diverse maize inbreds from Indian and CIMMYT research programmes were selected for screening for the presence of favourable alleles of lcyE and crtRB1 (Vignesh, Citation2012). The panel includes parents of released hybrids, elite inbreds, pre-breeding lines, specialty corns and inbreds newly developed from CIMMYT-HarvestPlus programme. Kernel colour of the selected inbreds varied from pale yellow to dark orange. Inbreds were grown at the ICAR-Indian Agricultural Research Institute (IARI) experimental farm, New Delhi, and were self pollinated. Individual ear in each line was harvested with husk and the grains were stored in darkness at 4°C until carotenoids extraction.
2.2. DNA extraction and PCR assay
DNA was isolated from young seedling of each inbred using standard CTAB procedure (Murray & Thompson, Citation1980) with minor modifications. PCR was performed using gene-specific markers for both the lcyE and crtRB1 genes. Primers for different genomic regions such as 5′TE of the lcyE gene and 3′TE of the crtRB1 gene were custom synthesized. We used the following set of primers, lcyE-5′TE-F: AAGCATCCGACCAAAATAACAG and lcyE-5′TE-R: GAGAGGGAGACGAC GAGACAC for lcyE 5′TE; crtRB1-3′TE-F: ACACCACATGGACAAGTTCG, crtRB1-3′TE-R1: ACACTCTGGCCCATGAACAC and crtRB1-3′TE-R2: ACAGCAATACAGGGG- ACCAG for the crtRB1 3′TE polymorphism. PCR amplification was carried out using the standard cycle conditions as given by Harjes et al. (Citation2008) and Yan et al. (Citation2010). Amplified fragments were separated using agarose gel electrophoresis and were scored for the presence of favourable alleles (Harjes et al., Citation2008; Yan et al., Citation2010).
2.3. Carotenoids extraction and quantification of kernel β-carotene
The carotenoids extraction and measurement were carried out as per the procedure described by Kurilich and Juvik (Citation1999) with minor modifications. A 20 μl volume of each sample was manually injected into the Water Alliance HPLC System (Waters Chromatography, Milford, MA) attached with a photodiode array detector (PDA). Six dilutions of β-carotene standard (SIGMA chemicals, USA) were used to make the standard curve and the concentration was detected by standard regression with external standard. The HPLC components include e2695 separation module and the 2996 PDA (Waters Corporation). The system was operated with Empower 2 Software (Waters Corporation). YMC carotenoid C30 column (5 μm, 4.6 × 250 mm; Waters Chromatography, Milford, MA) was used. The mobile phase consisted of methanol: tert-butyl methyl ether (80:20, v v−1) and the flow rate was 1 ml min−1. To maximize detection of carotenoids, absorbance was measured at 450 nm.
3. Results
PCR amplification of the diverse inbreds using gene-specific markers revealed polymorphisms for both lcyE (at 5′TE region) and crtRB1 (at 3′TE region) regulating carotenoid biosynthesis pathway.
3.1. Polymorphism for lycopene epsilon cyclase (lcyE) gene
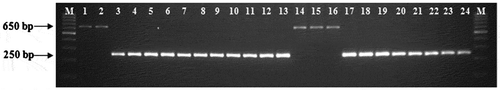
Among the inbreds screened in the study, 13 inbreds possessed 650 bp amplicon (favourable allele; allele 4) within the lcyE locus (Table ; Figure ). All other 372 inbreds in the panel amplified 250 bp amplicon (unfavourable allele; allele 2) in the 5′TE region of the gene. Among the 13 inbreds identified, two inbreds viz. HKI161 and HKI163 were from the Indian maize breeding programme. The other 10 inbreds viz. HP19-33, HP255-4, HP255-8, HP255-20, HP180-25, HP465-19, HP465-27, HP467-3, HP467-15 and HP467-20 belonged to CIMMYT-HarvestPlus programme (Table ). H16, an inbred (from CIMMYT-Africa) with white endosperm also possessed the favourable allele. The concentration of β-carotene in the yellow inbreds possessing favourable allele ranged from 0.19 to 11.51 μg g−1 with a mean of 4.45 μg g−1. The white maize inbred, H16, having recessive y1 allele (psy) did not possess any β-carotene in the kernel. Among the identified inbreds, only the five new set of HarvestPlus inbreds viz. HP465-19, HP465-27, HP467-3, HP467-15 and HP467-20 had higher concentration of β-carotene (8.61–11.51 μg g−1) and the rest of seven inbreds had less β-carotene (0.19–0.69 μg g−1) despite having the favourable allele.
Table 1. List of inbreds identified with the favourable allele (5′TE polymorphism) of lcyE gene (650 bp)
3.2. Polymorphism for crtRB1 gene
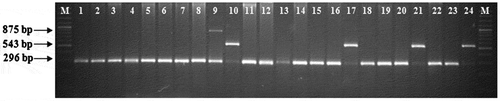
Among the 385 genotypes screened, 15 inbreds amplified 543 bp amplicon (favourable allele; allele 1) in the 3′TE region of the crtRB1 (Table ; Figure ). Among the inbreds identified with the favourable allele, SE547, Pant120, Pant125, V372, MGUDM-RIL47, KDMI4, EI116, MGUQPM-MAS4979 and BAJIM 06-10 are from the Indian maize breeding programme. The other inbreds viz. HP465-19, HP465-27, HP467-3, HP467-15 and HP467-20 belong to CIMMYT-HarvestPlus programme (Table ). Among the other inbreds that amplified the unfavourable alleles, 51 inbreds had allele 2 and 319 inbreds had allele 3. Estimation of β-carotene in the inbreds (having favourable allele) through HPLC revealed that 14 yellow inbreds had a mean β-carotene of 4.49 μg g−1 with a range of 0.41–11.51 μg g−1. Of the 15 inbreds identified in the study, surprisingly the inbred EI116 with white endosperm and absolutely no carotenoid also had the favourable allele for crtRB1 3′TE gene (Table ). Among the other 14 inbreds, only five in the new set of HarvestPlus inbreds viz. HP465-19, HP465-27, HP467-3, HP467-15 and HP467-20 had higher concentration of β-carotene (8.61–11.51 μg g−1) and the rest nine inbreds had less β-carotene (0.41–2.77 μg g−1), despite having the favourable allele. Of the 385 inbreds screened, only five inbreds viz. HP465-19, HP465-27, HP467-3, HP467-15 and HP467-20 had the favourable allele of both lcyE and crtRB1 genes.
Table 2. List of inbreds identified with the favourable allele (3′TE polymorphism) of crtRB1 gene (543 bp)
Figure 2. Allelic variations at 3′TE region of crtRB1 gene
4. Discussion
The present study analysed the presence of allelic diversity of lcyE (in 5′TE region) and crtRB1 (in 3′TE region) among diverse set of maize inbreds. In the present study, 3.38% of the inbreds had the favourable allele (allele 4) at lcyE locus, while 96.62% inbreds possessed the unfavourable allele (allele 2) (Table ). This is in congruence with the earlier report on the existence of the favourable allele in extremely less frequency (Harjes et al., Citation2008).
Though allele 4 is having the favourable effect, for most of the inbreds identified except for the new set of CIMMYT-HarvestPlus inbreds, the β-carotene in the endosperm was at par with the inbreds possessing the unfavourable allele (allele 2). This could be due to various reasons; firstly, carotenoid biosynthesis pathway is regulated by many genes, the identified inbreds may not be having the favourable alleles of other key genes governing enhancement of β-carotene in the pathway. This is well evident from the results of the present study that none of the eight inbreds (having low β-carotene) had the favourable allele of crtRB1, the other crucial gene responsible for higher accumulation of β-carotene. Even though the favourable lcyE allele diverts the pathway towards the β-branch, most of them will get hydroxylated to non-provitamin A carotenoids, like zeaxanthin, downstream in the pathway leading to drastic reduction of β-carotene in the endosperm (Vallabhaneni et al., Citation2009). In contrast, the new set of CIMMYT-HarvestPlus inbreds also had the favourable allele of crtRB1 gene and hence high β-carotene. Another interesting fact emerged was that H16, a white endosperm line showed the presence of lcyE 5′TE favourable allele. This inbred lacks the dominant allele for phytoene synthase (PSY1 or Y1), the gene which is responsible for the first committed step in the pathway. The dominant Y1 gene coverts geranylgeranyl diphosphate to phytoene and is responsible for the yellow colouration of the maize endosperm (Buckner et al., Citation1990). This phytoene serves as a substrate for further biochemical conversions in the pathway by the other downstream genes. Since the inbred H16 lacks the functional enzyme for this basic conversion step in the pathway, there may not be any substrate available for lcyE to act upon to divert them to β-branch of the pathway. Secondly, effect of the background genome in which this allele is present may also play a role in the regulation of carotenoid biosynthesis pathway. The favourable lcyE allele was originally identified in the temperate maize germplasm through association mapping approach (Harjes et al., Citation2008), whereas these alleles may not be effective in the genetic background of tropical and subtropical Indian maize inbreds. Babu et al. (Citation2013) also found similar results when they validated the effect of lcyE 5′TE allele across five populations. Thirdly, though the allele is amplifying 650 bp amplicon, there could also be some nucleotide polymorphisms present within the lcyE 5′TE region among the identified inbreds, leading to the variation in the β-carotene concentration.
The inbreds panel was also characterized for allelic variation in crtRB1 using gene-specific markers. Among the inbreds screened in the present study, 3.90% of the inbreds revealed the favourable allele (Figure ), while rest (96.10%) showed unfavourable alleles. The presence of the favourable allele in extremely less frequency in population supports earlier findings of Yan et al. (Citation2010).
CrtRB1 is a hydroxylase gene that causes hydroxylation of β-carotene to non-provitamin A carotenoids such as zeaxanthin. The favourable allele (543 bp) of crtRB1 gene causes reduced transcript expression, thereby blocking the conversion of β-carotene to zeaxanthin that leads to the higher accumulation of β-carotene in the maize kernel (Yan et al., Citation2010). However, among the identified inbreds having the favourable allele of crtRB1 3′TE gene, except for the new set of CIMMYT-HarvestPlus inbreds, other inbreds did not record high β-carotene concentration (0.41–2.77 μg g−1). The new set of CIMMYT-HarvestPlus genotypes having the favourable allele of crtRB1 3′TE gene showed a mean kernel β-carotene concentration of 10.1 μg g−1 (8.61–11.51 μg g−1).
The contrast in kernel β-carotene concentration could be attributed to the influence of genetic background in which they are present leading to difference in the phenotypic expression. This could also be due to the presence/absence of favourable alleles of other important genes in the carotenoid biosynthesis pathway. Inbreds (with favourable allele) with low β-carotene did not have the favourable allele of lcyE gene. Further to the results of lcyE gene, a white endosperm line EI116 showed favourable crtRB1 3′TE allele, but was devoid of any carotenoids. There could also be a possibility of nucleotide polymorphisms present in the crtRB1 gene that may cause difference in expression of the gene causing phenotypic variations (Vignesh, Nepolean, Hossain, Singh, & Gupta, Citation2013). This can be validated by comparing nucleotide difference vis-à-vis estimating the accumulation of transcript in the contrasting genotypes.
β-carotene-rich inbreds with favourable allele(s) of both lcyE and crtRB1 can be utilized in the breeding programme through many ways. Firstly, to develop provitamin A-rich maize hybrids, the new set of CIMMYT-HarvestPlus inbreds with high β-carotene and favourable alleles can be used as donors in the breeding programme (Vignesh et al., Citation2012). Since the effects of both genes are additive in nature (Yan et al., Citation2010), MAS can effectively introgress the favourable alleles of both crtRB1 and lcyE in diverse genetic backgrounds. Babu et al. (Citation2013) reported that the favourable crtRB1 allele is twice efficient in accumulating higher β-carotene when present in homozygous condition than under heterozygous condition. Thus, to develop hybrids homozygous for crtRB1 favourable allele, both the parents should be introgressed with the favourable allele. Secondly, newly developed CIMMYT-HarvestPlus inbreds (having high β-carotene) possess limited genetic diversity. Hence, these can be crossed with inbreds of Indian origin (having low β-carotene) to generate hybrids with moderate level of β-carotene. Thirdly, the favourable allele can be introgressed into the parents of already released hybrids through MAS to exploit the established grain heterosis and adaptability. Exploiting this strategy, two each of extra early and medium maturity hybrids have been improved for kernel β-carotene (Muthusamy et al., Citation2014).
Acknowledgements
Vignesh Muthusamy is thankful to the Indian Council of Agricultural Research (ICAR) for Senior Research Fellowship for his PhD programme. The authors thank Dr. Kevin Pixley (CIMMYT-Maize HarvestPlus Programme) for providing β-carotene-enriched lines used in the study. We also thank the breeders from AICMIP for sharing the inbred lines.
Additional information
Funding
Notes on contributors
Hari Shanker Gupta
ICAR-Indian Agricultural Research Institute, New Delhi, is a flagship organization for providing food and nutritional security in the country. Our research group pursue research for the alleviation of micronutrient malnutrition by biofortifying maize with micronutrients viz. provitamin A, vitamin E, Fe, Zn and essential amino acids viz. lysine and tryptophan through conventional and molecular breeding approaches. The research report in this paper aimed to identify maize inbreds with favourable alleles of two key genes viz. lycopene-ε-cyclase and β-carotene hydroxylase, governing higher accumulation of β-carotene, a major provitamin A carotenoid in maize kernel. These inbreds will serve as a rich genetic resource and can be utilized by breeding programmes for enrichment of β-carotene. The outcome of the study would help in developing biofortified maize hybrids to alleviate the widespread vitamin A deficiency in humans.
References
- Babu, R. , Rojas, N. P. , Gao, S. , Yan, J. , & Pixley, K. (2013). Validation of the effects of molecular marker polymorphisms in lcye and crtRB1 on provitamin A concentrations for 26 tropical maize populations. Theoretical and Applied Genetics , 126 , 389–399.10.1007/s00122-012-1987-3
- Black, R. E. , Allen, L. H. , Bhutta, Z. A. , Caulfield, L. E. , de Onis, M. , Ezzati, M. , … Maternal Child Under Nutrition Study Group . (2008). Maternal and child undernutrition: Global and regional exposures and health consequences. The Lancet , 371 , 243–260.10.1016/S0140-6736(07)61690-0
- Bouis, H. E. , Hotz, C. , McClafferty, B. , Meenakshi, J. V. , & Pfeiffer, W. H. (2011). Biofortification: A new tool to reduce micronutrient malnutrition. Food and Nutrition Bulletin , 32 , S31–40.
- Buckner, B. , Kelson, T. L. , & Robertson, D. S. (1990). Cloning of the y1 locus of maize, a gene involved in the biosynthesis of carotenoids. The Plant Cell Online , 2 , 867–876.10.1105/tpc.2.9.867
- Harjes, C. E. , Rocheford, T. R. , Bai, L. , Brutnell, T. P. , Kandianis, C. B. , Sowinski, S. G. , … Buckler, E. S. (2008). Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science , 319 , 330–333.10.1126/science.1150255
- Kurilich, A. , & Juvik, J. A. (1999). Quantification of carotenoid and tocopherol antioxidants in Zea mays . Journal of Agriculture and Food Chemistry , 47 , 1948–1955.
- Murray, M. G. , & Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research , 8 , 4321–4326.10.1093/nar/8.19.4321
- Muthusamy, V. , Hossain, F. , Thirunavukkarasu, N. , Choudhary, M. , Saha, S. , Bhat, J. S. , … Gupta, H. S. (2014). Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE , 9 , e113583.10.1371/journal.pone.0113583
- Vallabhaneni, R. , Gallagher, C. E. , Licciardello, N. , Cuttriss, A. J. , Quinlan, R. F. , & Wurtzel, E. T. (2009). Metabolite sorting of a germplasm collection reveals the hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiology , 151 , 1635–1645.10.1104/pp.109.145177
- Vignesh, M. (2012). Genetic analysis of provitamin A and marker-assisted breeding for β-carotene enrichment in maize ( PhD thesis). Indian Agricultural Research Institute, New Delhi.
- Vignesh, M. , Hossain, F. , Nepolean, T. , Saha, S. , Agrawal, P. K. , Guleria, S. K. , … Gupta, H. S. (2012). Genetic variability for kernel β-carotene and utilization of crtRB1 3′TE gene for biofortification in maize (Zea mays L.). Indian Journal of Genetics and Plant Breeding , 72 , 189–194.
- Vignesh, M. , Nepolean, T. , Hossain, F. , Singh, A. K. , & Gupta, H. S. (2013). Sequence variation in 3′UTR region of crtRB1 gene and its effect on β-carotene accumulation in maize kernel. Journal of Plant Biochemistry and Biotechnology , 22 , 401–408.10.1007/s13562-012-0168-4
- West, C. E. (2000). Vitamin A and measles. Nutrition Reviews , 58 , 546–554.
- Yan, J. , Kandianis, B. C. , Harjes, E. C. , Bai, L. , Kim, H. E. , Yang, X. , … Rocheford, T. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nature Genetics , 42 , 322–327.10.1038/ng.551
- Zhang, X. , Pfeiffer, W. H. , Palacios-Rojas, N. , Babu, R. , Bouis, H. , & Wang, J. (2012). Probability of success of breeding strategies for improving pro-vitamin A content in maize. Theoretical and Applied Genetics , 125 , 235–246.10.1007/s00122-012-1828-4