Abstract
SSR (simple sequence repeat) is a common molecular marker widely used in the analysis of rice genome and genetic variation research. While there have been some reports on factors affecting the quality of SSR molecular detection, no one has comprehensively explained these factors to date. Therefore, this study explores in detail the effects of electrophoresis buffer, electrophoresis time, and annealing temperature on the quality of SSR molecular detection in rice varieties. The results show that freshly prepared electrophoresis buffer, appropriate electrophoresis time, and suitable annealing temperature can improve the quality of SSR molecular detection in rice varieties and provide important technical support for in-depth research on the rice genome. Moreover, it is recommended that laboratories select and combine the aforementioned optimal experimental conditions to enhance the authenticity of rice varieties and improve the accuracy of molecular detection for purity.
PUBLIC INTEREST STATEMENT
The analysis of the rice genome and genetic variation research is of significant interest to the public. Simple Sequence Repeat (SSR) is a widely used molecular marker in this field. Despite some reports on factors affecting the quality of SSR molecular detection, a comprehensive explanation of these factors has been lacking. Therefore, this study aims to fill this gap by investigating the effects of electrophoresis buffer, electrophoresis time, and annealing temperature on the quality of SSR molecular detection in rice varieties. The findings reveal that the use of freshly prepared electrophoresis buffer, appropriate electrophoresis time, and suitable annealing temperature can enhance the quality of SSR molecular detection in rice varieties. These results provide crucial technical support for further research on the rice genome and contribute to the advancement of our understanding in this field, benefiting both scientific researchers and the general public.
1. Introduction
SSR (simple sequence repeat) is a commonly used molecular marker with high polymorphism and repeatability, which can be used for genetic variation and genome analysis of rice varieties (Kaur et al., Citation2015). Through the SSR molecular marker technology, the genetic similarity and diversity among rice varieties can be analyzed, providing important technical support for rice variety breeding, germplasm resource conservation, and variety identification (Cao et al., Citation2006; Jasim Aljumaili et al., Citation2018; Jayabalan et al., Citation2019).
In recent years, the Chinese Ministry of Agriculture has revised the rice variety identification standard, issuing the national standard GB/T 39,917–2021. This standard outlines the technical requirements and methods for rice variety identification, with SSR molecular marker technology being recognized as an essential tool for this purpose (Ashraf et al., Citation2016; Yang et al., Citation2021; Zhou et al., Citation2023). However, the reliability and accuracy of molecular detection results depend on experimental conditions and operator skills. To enhance the accuracy of identifying the authenticity and purity of rice varieties, this study focuses on three specific rice varieties: the sterile lines Hui Xiang A and Ba Xiang A, the hybrid rice Te You 816, and the conventional rice Meiya silk. Individual loci were assessed using a pair of SSR primers, with each polymorphic band representing an allele (Chen et al., Citation2017). Primers corresponding to the male sterile lines of Hui Xiang A and Ba Xiang A predominantly produced a single allele, making them suitable for investigating the impact of the electrophoresis buffer on assay quality (Lee et al., Citation2012). The hybrid rice Te You 816 exhibited well-defined alleles corresponding to the primers, providing an opportunity to determine the optimal electrophoresis time (Azqueta et al., Citation2011). The conventional rice Meiya silk displayed a higher number of polymorphic genes, prompting the exploration of different annealing temperatures and their effect on detection quality. By optimizing these experimental conditions, the study aims to improve the accuracy and repeatability of SSR molecular detection, thereby providing crucial technical support for in-depth research on the rice genome.
2. Materials and methods
2.1. Materials
The experimental materials used in this study were rice varieties provided by Guangdong Tianlian Seed Industry Co., Ltd., including Hui Xiang A and Ba Xiang A (male sterility lines), Te You 816 (hybrid rice), and Meiya silk (conventional rice).
2.2. Primers
48 pairs of SSR primers were selected from the GB/T 39,917–2021: SSR molecular marker detection for the authenticity and purity of major crops—Rice (implemented on 1 November 2021). The primer names and sequences are detailed in Table , and the primers were synthesized by Generay Biotechnology.
Table 1. Rice identification primers
2.3. Reagents
DNA extraction: TianGen Plant Genomic DNA Extraction Kit (DP305) from TianGen Biotech Co., Ltd. was used for rice DNA extraction.
PCR amplification system: 2X EasyTaq PCR SuperMix (AS111) from TRANS.
Electrophoresis: Agarose from IOWEST, 5XTBE Buffer from Yanoon Biotech Co., Ltd., nucleic acid dye (GS101) from TRANS, GeneRuler 100bp DNA Ladder (SM0241) from Thermo Fisher Scientific.
2.4. Main instruments
DNA concentration detection: NanoDrop 1000 ultraviolet spectrophotometer from Thermo Fisher Scientific.
PCR amplification: Bio-Rad T100 was used for PCR amplification.
Electrophoresis: Beijing Liuyi DYCP-31DN equipment.
Gel imaging: Vilber Lourmat FUSIONFX7 imaging system from France was used for agarose gel exposure and photography.
2.5. Experimental design
In this experiment, electrophoresis was performed using electrophoresis buffer that was reused more than three times and freshly prepared 0.5XTBE electrophoresis buffer, and the clear differences between the exposed bands before and after were compared. After electrophoresis at a constant voltage of 140 V, the separation of exposed bands after 40 min, 50 min, and 75 min of electrophoresis was compared to observe the differences in electrophoresis results. PCR amplification was performed on the DNA template at three annealing temperatures of 55°C, 60°C, and 67°C to compare the specificity of the PCR products.
3. Results
3.1. DNA quality detection
DNA was extracted using a DNA extraction kit, and the results of the ultraviolet spectrophotometer detection were shown in Table . The absorption wavelength of 260 nm is the highest absorption peak of nucleic acids, while 280 nm is the absorption peak of proteins often used to determine the purity of the sample. For pure DNA, the A260/A280 ratio should be between 1.7 and 1.9, and for pure nucleic acid, the A260/A230 ratio is generally between 1.8 and 2.0. The results showed that the purity of the extracted DNA met the standard and could be used for the next experiment.
Table 2. Rice DNA concentration
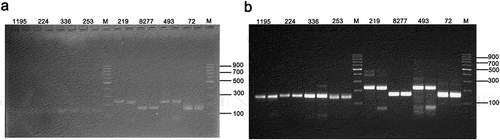
3.2. Impact of the quality of electrophoresis buffer on the detection of variety authenticity using SSR molecular markers
In this study, we extracted DNA from the rice varieties of Hui Xiang A seedling and Ba Xiang A seedling, and compared the impact of using electrophoresis buffer that had been used for at least three runs (Figure ) versus freshly prepared electrophoresis buffer (Figure ) on the detection of variety authenticity using SSR molecular markers. The results showed that when using freshly prepared electrophoresis buffer for analysis, the bands produced were significantly clearer and easier to distinguish, indicating a significant difference between the two methods.
Each primer pair was run in two wells, with Hui Xiang A seedlings in the first well and Ba Xiang A seedlings in the second well. (a) Results obtained using electrophoresis buffer that had undergone at least three runs. (b) Results obtained using freshly prepared electrophoresis buffer.
3.3. Impact of electrophoresis time on the detection of variety authenticity using SSR molecular markers
Preliminary experiments were conducted to assess the influence of electrophoresis time on the authenticity detection of SSR molecular marker varieties. The test variety chosen was Te You 816, a recognized high-yielding rice variety from the gene bank. The National Crop Variety DNA Fingerprint Database Platform (https://202.127.44.15/) was utilized to select representative primer templates for PCR amplification, including the one-allelic site RM253 and the two-allelic site RM72. Through analysis of the experimental data, we found that increasing the electrophoresis time appropriately can improve the accuracy of results for shorter amplified lengths. At 40 minutes (Figure ) and 50 minutes (Figure ) of electrophoresis, the RM72 primer produced a single, thick band. However, after 75 minutes of electrophoresis (Figure ), the number of bands for RM72 became double while RM253 remained a single band, which matched the results in the fingerprint library. After 90 minutes of electrophoresis, the band ran out of the gel because the electrophoresis time was too long, and the band disappeared, so it was not recorded. This indicates that electrophoresis time has a significant impact on the results of the detection of variety authenticity using SSR molecular markers in this clinical trial.
Figure 2. Impact of electrophoresis time on the detection of variety authenticity using SSR molecular markers.
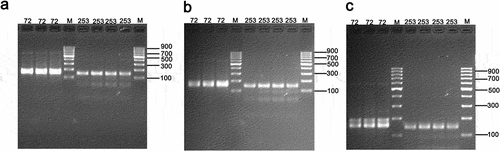
Extract DNA from the Te You 816 and amplify the RM253 and RM72 sequences at different electrophoresis times (40 minutes, 50 minutes, 75 minutes). (a) Results of electrophoresis after 40 minutes of PCR; (b) Results of electrophoresis after 50 minutes of PCR; (c) Results of electrophoresis after 75 minutes of PCR. (n = 3).
3.4. Impact of annealing temperature on the detection of variety authenticity using SSR molecular markers
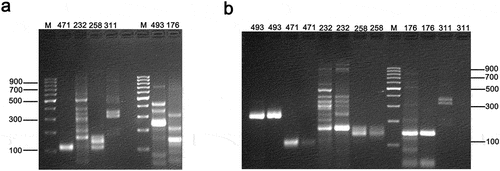
In this experiment, DNA was extracted from Meiya silk, and 48 primer pairs were amplified at 94°C for denaturation, 55°C for annealing, and 72°C for extension. After electrophoresis, we found that multiple bands appeared for some primers, leading us to suspect that these bands might be non-specific (Supplementary Figure S1). To address this issue, we selected several primers and conducted gradient PCR detection at different annealing temperatures. According to the national standard GB/T 39,917–2021, the recommended annealing temperature is 55°C for most samples, except for RM119 and RM176, which require annealing at 67°C. Therefore, an additional intermediate temperature of 60°C was included. The aim was to observe the impact of different annealing temperatures on the amplification results.
From the experimental results, we found that the RM176 primer produced the best PCR amplification results at an annealing temperature of 67°C (Figure ), without producing any additional non-specific bands. For the RM493 primer, higher annealing temperatures of 60°C or 67°C (Figure ) significantly prevented the appearance of non-specific bands, producing only a single band. The RM311 primer achieved specific double band amplification at an annealing temperature of 60°C (Figure ). The RM471 primer achieved specific single-band amplification at an annealing temperature of 55°C (Figure ). However, the RM232 primer did not eliminate non-specific bands at three different annealing temperatures, requiring further research and exploration. Therefore, appropriate annealing temperatures must be identified for different primers to obtain specific bands.
Extract DNA from the Te You 816 and amplify the sequences at different annealing temperatures (55°C, 60°C, 67°C). (a): PCR product performance at an annealing temperature of 55°C; (b): Same primer, with the first well at an annealing temperature of 60°C and the second well at an annealing temperature of 67°C.
4. Discussion
The SSR method is widely used in rice varieties for genetic diversity analysis, marker-assisted breeding, and genomic mapping (Kaur et al., Citation2015). However, the accuracy of SSR will be affected by many factors, including the quality of the extracted DNA, the amplification procedure of PCR and the separation of amplification products, etc.
The quality of DNA extraction is a critical factor that can affect the quality of SSR molecular detection. High-quality DNA extraction is essential to ensure that the PCR amplification and electrophoresis result in reliable and accurate SSR data.
We use a low-cost, reproducible, and easy-to-operate agarose gel electrophoresis method to separate the amplified products (Azqueta et al., Citation2011; Yao et al., Citation2013). Electrophoresis buffer plays a key role in protein separation and migration during electrophoresis. The ion concentration and pH value in the electrophoresis buffer will affect the clarity and accuracy of the electrophoresis results (Hu et al., Citation2020). Therefore, we found that fresh electrophoresis buffer can make the agarose gel run clearer, thus improving the resolution of SSR bands and accurate genotyping. Therefore, regular preparation and replacement of electrophoresis buffer is necessary.
Furthermore, we have observed a significant influence of electrophoresis time on the detection results of SSR molecular markers for varietal authenticity. Previous studies have reported electrophoresis times of 40 minutes for PCR product detection (O’Conner et al., Citation1991), while other studies have reported electrophoresis times of 90 minutes for PCR product detection (Liu et al., Citation2021). Based on our laboratory experience, we conducted electrophoresis for durations of 40 minutes, 50 minutes, 75 minutes, and 90 minutes, followed by gel imaging system identification. Through experimentation, we found variations in the number and intensity of SSR bands produced by the same batch of rice samples under different electrophoresis times. The optimal electrophoresis time determined in this experiment was 75 minutes. This indicates the need to optimize the electrophoresis time according to specific conditions in SSR analysis to obtain more accurate and stable results.
The annealing temperature is one of the parameters that affect the purity and yield of reaction products. Unsuitable annealing temperatures can lead to the formation of non-specific products and decrease product yield (Wang, Citation2006). This aligns with the findings of various other studies, such as the research conducted by K. Ishii et al (Rychlik et al., Citation1990), which demonstrated that optimizing the annealing temperature reduces the impact of primer mismatch in PCR. In the context of rice SSR analysis, Yu Xiaoyue (Ishii & Fukui, Citation2001) confirmed that annealing at 55°C is beneficial for detecting the authenticity of SSR markers in rice varieties. We also validated that most PCR products annealed at 55°C yielded the best results. However, we observed that the primers RM176 and RM493 effectively eliminated the influence of non-specific bands at an annealing temperature of 67°C, while RM311 achieved the same outcome at 60°C. Therefore, determining the appropriate annealing temperature for each primer is essential to ensure the accuracy and stability of SSR molecular marker detection results.
While the abovementioned factors have been investigated and established as important, other factors that can affect the quality of SSR molecular detection need to be further explored. For instance, the effects of PCR cycle number (Xiaoyue et al., Citation2016), primer concentration (Killgore et al., Citation2000), and DNA template concentration (Gunson et al., Citation2003) on SSR data quality should be investigated in future studies.
In summary, this study systematically explored and analyzed the impact of experimental parameters such as electrophoresis buffer, electrophoresis time, and annealing temperature on the detection of rice variety authenticity using SSR molecular markers. The research results provide important reference for the design and implementation of experiments, and provide new ideas and methods for related research. However, this study still has shortcomings and requires further in-depth exploration and verification.
5. Conclusion
The SSR marker method has gained wide acceptance for authenticating rice varieties due to its rapid and stable characteristics. This article aims to explore the seamless execution of detection work by selecting optimal PCR reaction conditions and electrophoresis parameters tailored to the laboratory’s specific requirements. In the process of utilizing the SSR method to determine rice variety authenticity, a critical step lies in carefully controlling the annealing temperature during PCR amplification. Only by amplifying specific and low-impurity products, subsequently performing electrophoresis in fresh electrophoresis solution, and choosing the appropriate electrophoresis duration, can distinct bands and a pristine background fingerprint pattern be achieved, thereby facilitating subsequent reading, analysis, and identification. These findings lay a robust research foundation for employing SSR marker technology in rice variety authentication.
Supplemental Material
Download ()Acknowledgments
We thank the Laboratory Department of the First Affiliated Hospital of Sun Yat-sen University for financial support and Guangdong Tianlian Seed Industry Co., Ltd. for providing materials. Special thanks to Zheng Liang and Jingrou Chen for their experimental work, data analysis, Chenglong Pan and Yang Wang for image editing. We also appreciate the guidance from Zongjun Zhang, Min Liu, and Peisong Chen in preparing this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/23311932.2023.2275414
Additional information
Notes on contributors

Peisong Chen
Jingrou Chen, a medical master’s graduate and a laboratory physician in the Department of Clinical Laboratory at the First Affiliated Hospital of Sun Yat-sen University. Her research focuses on stem cells, immunology, molecular diagnostics of diseases, and molecular diagnostics in agriculture. She has participated in one project funded by the National Natural Science Foundation and one provincial-level project. She has published eight papers, six of which are indexed in SCI, with two as the first author/co-author. Additionally, she holds one patent.

Jingrou Chen
Jingrou Chen, a medical master’s graduate and a laboratory physician in the Department of Clinical Laboratory at the First Affiliated Hospital of Sun Yat-sen University. Her research focuses on stem cells, immunology, molecular diagnostics of diseases, and molecular diagnostics in agriculture. She has participated in one project funded by the National Natural Science Foundation and one provincial-level project. She has published eight papers, six of which are indexed in SCI, with two as the first author/co-author. Additionally, she holds one patent.
References
- Ashraf, H., Husaini, A. M., Ashraf Bhat, M., Parray, G. A., Khan, S., & Ganai, N. A. (2016, October). SSR based genetic diversity of pigmented and aromatic rice (Oryzasativa L.) genotypes of the western Himalayan region of India. Physiology and Molecular Biology of Plants., 22(4), 547–9. https://doi.org/10.1007/s12298-016-0377-8
- Azqueta, A., Gutzkow, K. B., & Brunborg, G. (2011, September 18). Towards a more reliable comet assay: Optimising agarose concentration, unwinding time and electrophoresis conditions. Mutation Research, 724(1–2), 41–45. https://doi.org/10.1016/j.mrgentox.2011.05.010
- Cao, Q., Lu, B. R., Xia, H., RONG, J., SALA, F., SPADA, A., & GRASSI, F. (2006, December). Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in North-eastern China revealed by simple sequence repeat (SSR) markers. Annals of Botany, 98(6), 1241–1252. https://doi.org/10.1093/aob/mcl210
- Chen, J., Xie, W., & Wang, T. (2017). Genetic diversity analysis of photoperiod and temperature-sensitive male sterile lines in rice based on SSR markers. Jiangxi Journal of Agricultural Sciences, 29(3), 1–6.
- Gunson, R., Gillespie, G., & Carman, W. F. (2003, April). Optimisation of PCR reactions using primer chessboarding. Journal of Clinical Virology, 26(3), 369–373. https://doi.org/10.1016/S1386-6532(03)00006-4
- Hu, W., Zhou, T., Wang, P., Wang, B., Song, J., Han, Z., Chen, L., Liu, K., & Xing, Y. (2020, January). Development of whole-genome agarose-resolvable LInDel markers in rice. Rice (N Y), 13(1), 1. https://doi.org/10.1186/s12284-019-0361-3
- Ishii, K., & Fukui, M. (2001, August). Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Applied & Environmental Microbiology, 67(8), 3753–3755. https://doi.org/10.1128/AEM.67.8.3753-3755.2001
- Jasim Aljumaili, S., Rafii, M. Y., Latif, M. A., Sakimin, S. Z., Arolu, I. W., & Miah, G. (2018, March 15). Genetic diversity of aromatic rice germplasm revealed by SSR markers. BioMed Research International, 2018, 1–11. https://doi.org/10.1155/2018/7658032
- Jayabalan, S., Pulipati, S., Ramasamy, K., Jaganathan, D., Venkatesan, S. D., Vijay, G., Kumari, K., Raju, K., Hariharan, G. N., & Venkataraman, G. (2019, September 10). Analysis of genetic diversity and population structure using SSR markers and validation of a Cleavage Amplified Polymorphic Sequences (CAPS) marker involving the sodium transporter OsHKT1;5 in saline tolerant rice (Oryza sativa L.) landraces. Gene, 713, 143976. https://doi.org/10.1016/j.gene.2019.143976
- Kaur, S., Panesar, P. S., Bera, M. B., & Kaur, V. (2015). Simple sequence repeat markers in genetic divergence and marker-assisted selection of rice cultivars: A review. Critical Reviews in Food Science and Nutrition, 55(1), 41–49. https://doi.org/10.1080/10408398.2011.646363
- Killgore, G. E., Holloway, B., & Tenover, F. C. (2000, July). A 5’ nuclease PCR (TaqMan) high-throughput assay for detection of the mecA gene in staphylococci. Journal of Clinical Microbiology, 38(7), 2516–2519. https://doi.org/10.1128/JCM.38.7.2516-2519.2000
- Lee, P. Y., Costumbrado, J., Hsu, C. Y., & Kim, Y. H. (2012, April). Agarose gel electrophoresis for the separation of DNA fragments. Journal of Visualized Experiments: JoVe, 20(62), 3923. https://doi.org/10.3791/3923
- Liu, Q., Ding, X., & Zhang, P. (2021). Purity identification of hybrid rice Seed using SSR molecular markers. Anhui Agricultural Bulletin, 27(5), 9–10.
- O’Conner, J. L., Wade, M. F., & Zhou, Y. (1991, March). Control of buffer pH during agarose gel electrophoresis of glyoxylated RNA. Biotechniques, 10(3), 300–302.
- Rychlik, W., Spencer, W. J., & Rhoads, R. E. (1990, November). Optimization of the annealing temperature for DNA amplification in vitro;. Nucleic Acids Research, 18(21), 6409–6412. https://doi.org/10.1093/nar/18.21.6409
- Wang, X. (2006). Construction of rice characteristic fingerprint and analysis of seed purity using SSR markers. Fujian Agriculture and Forestry University.
- Xiaoyue, Y., Hua, Y., & Fanggang, Z. (2016). Effect of annealing temperature, DNA polymerase, and thermal cycler on the quality of rice variety authenticity SSR molecular marker detection. Jiangsu Agricultural Sciences, 44(11), 57–60.
- Yang, G., Yang, Y., Guan, Y., Xu, Z., Wang, J., Yun, Y., Yan, X., & Tang, Q. (2021, October 19). Genetic diversity of Shanlan Upland rice (Oryza sativa L.) and association analysis of SSR markers linked to agronomic traits. BioMed Research International, 2021, 1–11. https://doi.org/10.1155/2021/7588652
- Yao, G., Lu, L., & Dan, Z. (2013). Screening and evaluation of agarose gel electrophoresis polymorphic SSR markers between indica and japonica subspecies. Henan Agricultural Sciences, 42(08), 16–20. https://doi.org/10.15933/j.cnki.1004-3268.2013.08.030
- Zhou, H., Li, S., Liu, J., Hu, J., Le, S., & Li, M. (2023, February). Identification and analysis of the genetic integrity of different types of rice resources through SSR markers. Scientific Reports, 13(1), 2428. https://doi.org/10.1038/s41598-023-29514-y