Abstract
To characterize field strains of Mycoplasma gallisepticum (MG) in Malaysia, selected gene target-specific sequences to MG, hemagglutinin protein A (pMGA) and phase variable putative adhesion protein A (pvpA) genes, were amplified from positive MG samples using conventional polymerase chain reaction (PCR). A total of 25 MG-positive samples out of 94 field samples were sequenced with primer targeting pMGA and pvpA genes. Sequencing and phylogenetic analysis were conducted using bioinformatics softwares (Bioedit and MEGA 5) and genetic variation patterns were evaluated based on partial nucleotide sequencing of pMGA and pvpA genes. In case of pMGA, all 25 field strains showed similar gene size patterns with strains of pathogenic reference (MGS6) and vaccine (MG F), which were different from the PCR product size of less pathogenic vaccine strain (TS 11). However, pvpA partial nucleotide sequences of MG local strains showed that 20 out of 25 strains possessed similar gene size pattern with MG F, which were different from MGS6 and TS 11. In conclusion, based on the molecular characterization from phylogenetic analysis of pMGA and pvpA partial nucleotide sequences, it was found that Malaysian MG strains were different from strains reported in other countries.
Public Interest Statement
Mycoplasma gallisepticum is a primary pathogen that can cause chronic respiratory disease (CRD) which leads to severe economic losses to the commercial poultry industry through reduced feed efficiency, decreased egg production and increased mortality. Although the farmers follow strict biosecurity management, vaccination programme and medicinal treatment in the poultry farm to prevent this disease, recurrent infection and outbreaks are still present. Therefore, the control of mycoplasmosis is very important in order to increase the output of poultry farm. MG genes are related with pathogenic, antigenic and immune evasion properties. It has been reported that molecular characterization enables epidemiological tracing and classification of new field MG strains. So, this study will be a core for further efficient evaluation to develop a potential strategy for prevention and control of MG infection.
Competing interests
The authors declare no competing interests.
1. Introduction
Mycoplasma are members of the Mycoplasmataceae family, which belongs to the Mycoplasmatales order in the Mollicutes class (Sasaki et al., Citation2002); and there are more than 200 species in this class. One of the species is Mycoplasma gallisepticum (MG), which has a worldwide distribution and is the most economically important pathogenic avian Mycoplasma. MG is a primary pathogen that can cause chronic respiratory disease (CRD) which leads to severe economic losses to the commercial poultry industry (Liu, Garcia, Levisohn, Yogev, & Kleven, Citation2001).
Sequencing methods have been introduced to study molecular epidemiology of bacterial pathogens (Enright & Spratt, Citation1999). To differentiate MG strains, the improvement on the molecular biology of MG (Razin, Yogev, & Naot, Citation1998) and MG complete genome sequence availability have been used to evaluate gene-targeted sequencing as a typing tool (Papazisi et al., Citation2003). DNA polymorphisms are the basis for the application of PCR based methods to detect and differentiate MG strains. Nascimento, Yamamoto, Herrick, and Tait (Citation1991) showed that, gene-targeted sequencing analysis might help in the synthesis of specific strain primers and can be used to detect MG strains.
The adherence of MG can be affected by the changes in expression of both pMGA and cytadhesin genes (mgc1 and mgc3) but pMGA genes play a big potential role for antigenic variants generation (Bencina, Citation2002). Basically, pMGA and pvpA are two gene families of MG that encode major surface proteins having pathogenic, antigenic and immune evasion properties (Markham, Glew, Whithear, & Walker, Citation1993). The pMGA gene family involves a minimal of 16% of R strain and 7.7% of F strain (Baseggio, Glew, Markham, Whithear, & Browning, Citation1996) which are powerful genomic engagements to antigenic variability, and hypothetically play a key role in immune evasion (Markham et al., Citation1993). The pMGA gene family also accommodate a system for prompt and reversible switches in its expression of proteins (antigenic switching) in response to antibodies or other environmental clues (Glew, Browning, Markham, & Walker, Citation2000). The pvpA is integral membrane proteins of MG having size variation that possess high-frequency phase and antigenic capability (Levisohn, Rosengarten, & Yogev, Citation1995). In this study, genes used for sequencing were pMGA and pvpA which had size polymorphism (Tan, Citation2008).
In Malaysia, MG field strains showed gene size polymorphism in pMGA and pvpA genes with a significant pattern in comparison with reference pathogenic and vaccine strains (Zahraa, Aini, Omar, Hair-Bejo, & Tan, Citation2011). However, there is very limited information on molecular characterization; regarding the comparison of Malaysian local strains with international strains, upon analysis of phylogenetic tree based on pMGA and pvpA genes. Therefore, the objectives of the study were to sequence pMGA and pvpA genes of MG field strains and also to characterize these local strains based on pMGA and pvpA phylogenetic analysis.
2. Materials and methods
2.1. Mycoplasma gallisepticum samples
A total of 300 swab samples were obtained from different states of poultry farms in Malaysia where 94 were MG-positives field strains by real-time PCR using gapA 5F + 6R primer set of gapA gene (Yasmin et al., Citation2014). Ninety-four positive samples were also amplified by conventional PCR using AUTS11 F + ATTS11 R and pvpA 1F + pvpA 2R primer sets of pMGA and pvpA genes accordingly. Based on the brightness and thickness of bands, 25 field strains out of 94 samples were selected and amplified again by conventional PCR for molecular characterization. The sampling size, location, vaccination, farming type and number of positive samples are summarized in Table .
Table 1. Sampling size, sampling location and vaccination history of layer breeder, broiler breeder, broiler chicken and village chicken used in this study
2.2. Extraction of genomic DNA
An ethanol-cleaned forceps was used each time to remove cotton swab from the phosphate-buffered saline (PBS) under sterile condition. Conventional salt-based method was used to extract genomic DNA with some modifications. In the first step of DNA extraction, 100 μl of 10% sodium dodecyl sulphate (SDS) was used to lyse cells and tissues as well as to remove lipid membrane. Then, cellular and histone proteins bound to DNA were removed by adding 2 μl of 50 μg/μl Proteinase K (Epicentre® Biotechnologies, USA). The pellet was resuspended by vortexing the mixture. The mixture was then incubated at 65°C for 30 min and thoroughly shaked in every 5 min. Then, all the tubes were left at −20°C for 8 min or at 4°C for 30 min to 1 h.
Protein and peptide were precipitated by adding 300 μl of ammonium acetate and vortex for 10 s to mix it properly. A micropipette was used to resuspend the pellet if it stuck at the bottom of tube and the pellet was centrifuged at 14,000 rpm for 15 min. Then, a volume of 850 μl from the supernatant was saved by transferring it into another new microcentrifuge tube. For precipitation of DNA, a volume of 550 μl of isopropanol was added into the supernatant and the tube was inverted about 40 times. The mixture was then centrifuged at 14,000 rpm for 15 min at 4°C. The isopropanol was then poured off carefully to avoid DNA pellet dislodging. Resultant DNA pellet was washed twice in 1 ml of 75% (v/v) ethanol by centrifugation at14,000 rpm for 15 min at 4°C. The ethanol was poured off carefully so that no dislodge of DNA pellet. The DNA samples were then placed as inverted position in the laminar air flow chamber for 2 h until the evaporation of ethanol. At last, these samples were resuspended in 30 μl of nuclease free water, then kept at 4°C for 30 min and finally stored at -20°C until to be used for PCR.
2.3. PCR assay targeting pMGA and pvpA genes
For molecular characterization, field strains of positive MG with reference strains were amplified by conventional PCR for gene sequencing. Primers used in this study (Table ), were synthesized by First Base Laboratories Sdn. Bhd., Selangor, Malaysia. The PCR protocol was optimized by varying conditions, viz. DNA concentration, amplification cycle, amplification time and primer annealing temperature. An automatic thermal cycler (MyCycler, BioRad, USA) was used to amplify fragmented DNA and the reaction set-up was consisted of an initial denaturation step at 94°C for 4 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 52°C (for pMGA gene) and 58°C (for pvpA gene) for 60 s, extension at 72°C for 15 s and final extension at 72°C for 5 min. The reaction volume was set up in a 25-μl reaction mixture where 12.5 μl (2X) (Bioline, Malaysia) of 2X MyFiMix, 0.4 μl (25 pmol) of forward primer, 0.4 μl (25 pmol) of reverse primer, 2 μl (200 ng) of DNA template and 9.7 μl of distilled water. The gel electrophoresis system at 1.5% agarose gel was used to view the result of amplified DNA in 1X TAE performed at 90 V for 40 min, stained with ethidium bromide (10 mg/ml). Bands were visualized on UV light and the photograph was taken with analyser (Syngene, Gene Genius Bioimaging System). PCR products were compared with 100-bp plus DNA ladder (Vivantis®, Malaysia) and the presence of band indicated a positive result, whereas the absence of band indicated a negative result.
Table 2. PCR product size and primer sequences of pMGA and pvpA genes used for MG characterization
2.4. DNA purification, DNA sequencing and sequence analysis
Positive bands were selected for DNA purification which was performed using the PCR SV kit (Gene All® ExpinTM PCR SV). Purified DNA of PCR products were sent for sequencing to Macrogen Inc, Seoul, Korea. Nucleotide sequences of pMGA and pvpA genes were identified through direct sequencing from both directions using primers as shown in Table . The Automatic Sequencer-ABI 3730Xl DNA Analyzer using the ABI PRISM® BigDyeTM Terminator Cycle Sequencing Kits with AmpliTaq DNA Polymerase (FS enzyme) (Applied Biosystem, CA, USA) was used to perform sequencing reactions. Purified DNAs of reference pathogenic (MGS6, MG R and PG 31) and vaccine (TS 11, MG F and 6/85) strains were also sent for sequencing. The web-based pair-wise BLAST (Basic Local Alignment Search Tool) search programs under NCBI (National Centre for Biotechnology Information) was used for the primary determination and comparison among different strains of the raw sequence data. The Bioedit Sequence Alignment Editor Program with 7.0.9.0 version (Hall, Citation1999); based on Clustal W Multiple alignment (Thompson, Higgins, Gibson, & Clustal, Citation1994); was used for the assembling and analysing of sequence data. This program was also used to find sequence identity matrix. Multiple sequence alignment of local field strains, reference strains and international sequences were also analysed using the MEGA with version 5 software program (Tamura et al., Citation2011). MG international sequences were collected from Gene Bank (Table ).
Table 3. MG International sequences used for sequence alignment and phylogenetic analysis
2.5. Phylogenetic analysis
To fulfil the phylogenetic analysis, MG field strains together with reference strains were compared with strains of other countries as shown in Table . The neighbor joining tree of the MEGA with version 5 software was used to do phylogenetic analysis (Tamura et al., Citation2011). The accuracy of clustering of the neighbor-joining tree was assessed using 1000 bootstrap replicates.
3. Results
3.1. Amplification of MG field strains by conventional PCR
For sequencing, 25 positive samples were selected for each pMGA and pvpA gene, which were based on the brightness and thickness of bands (Figures –). Both pMGA and pvpA genes had size polymorphism on specific target sequence. Specific primer targeting pMGA and pvpA genes could detect the specific gene on the MG polymerized reaction with the primer-specific annealing temperature, and thus were able to detect the presence of MG in field samples.
Figure 1. Picture of PCR products of MG reference and field strains (12 field samples) by pMGA gene using primer AUTS11 F + ATTS11 R. 100-bp plus DNA ladder (lane 1), MGS6 (lane 2), MG F (lane 3), TS 11 (lane 4), 6/85 (lane 5), MG R (lane 6), PG 31 (lane 7), A5969 (lane 8), MG-positive field strains (lane 9–20), Negative control (lane 21).
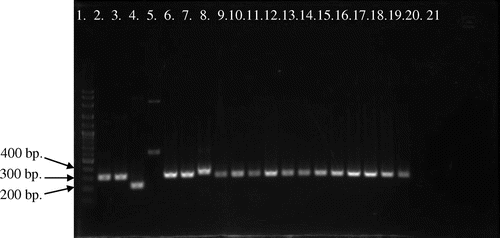
Figure 2. Picture of PCR products of MG reference strains and local strains (13 field samples) by pMGA gene using primer AUTS11 F + ATTS11 R. 100-bp plus DNA ladder (lane 1), MGS6 (lane 2), MG F (lane 3), TS 11 (lane 4), 6/85 (lane 5), MG R (lane 6), PG 31 (lane 7), A5969 (lane 8), MG-positive field strains (lane 9–21), Negative control (lane 22).
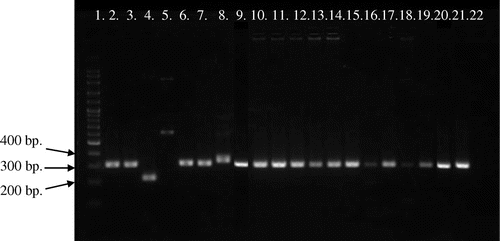
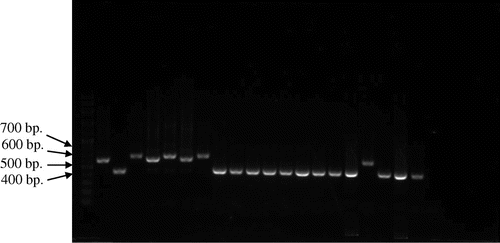
Figure 3. Picture of PCR products of MG reference strains and local strains (12 field samples) by pvpA gene using primer pvpA 1F + pvpA 2R. 100-bp plus DNA ladder (lane 1), MGS6 (lane 2), MG F (lane 3), TS 11 (lane 4), 6/85 (lane 5), MG R (lane 6), PG 31 (lane 7), A5969 (lane 8), MG-positive field strains (lane 9–20), Negative control (lane 21).
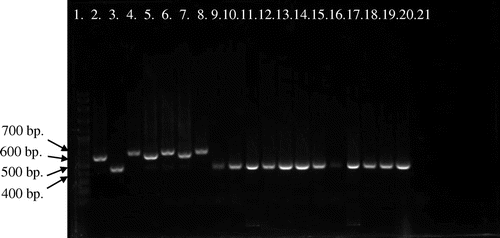
Figure 4. Picture of PCR products of MG reference strains and local strains (13 field strains) by pvpA gene using primer pvpA 1F + pvpA 2R. 100-bp plus DNA ladder (lane 1), MGS6 (lane 2), MG F (lane 3), TS 11 (lane 4), 6/85 (lane 5), MG R (lane 6), PG 31 (lane 7), A5969 (lane 8), MG-positive field strains (lane 9–21), Negative control (lane 22).
3.2. PCR result of MG field strains by pMGA gene
Analysis of MG PCR products of the pMGA gene (or M9 gene) by 1.5% agarose gel electrophoresis indicated that the location of bands were within 200–400 bp (Figures and ). Gene size variations were observed among PCR products of the pMGA gene both local field and reference strains. PCR product sizes of field strains used in this study were varied in between 315 and 336 bp, whereas product sizes of reference strains were ranged in between 226–329 bp. So, the overall range of PCR product sizes were 226–336 bp when were amplified by AUTS11 F + ATTS11 R primer set of pMGA gene. The smallest product length observed in vaccine strain (TS 11) was 226 bp, whereas the largest product found in MM33 field strain was 336 bp. Moreover, rest of 24 field strains showed similar gene size pattern with reference pathogenic (MGS6) and virulence vaccine (MG F) strains which were different from the less virulence vaccine strain TS 11 (PCR product 226 bp) and the reference strain A5969 (PCR product 329 bp). However, although all strains showed single band, the vaccine strain (6/85) showed multiple bands indicating nonspecific to the primer set of pMGA gene.
3.3. PCR result of MG field strains by pvpA Gene
Analysis of MG PCR product size of pvpA gene by 1.5% agarose gel electrophoresis indicated that the location of bands were within 400–700 bp (Figures and ). Gene size variations were observed within PCR products of the pvpA gene among field and reference strains. This study also showed the gene size polymorphism among PCR products of MG local strains. PCR product sizes of local and reference strains were varied in between 434–665 bp and 446–677 bp, respectively. So, the overall range of PCR products were ranged in between 434 and 677 bp when were amplified by pvpA 1F + pvpA 2R primer set of pvpA gene. The smallest product length was 434 bp, which was observed in local strains of MM8, MM10, MM12, MM14 and MM19, whereas the largest product size was 677 bp found in the reference strain MG R.
3.4. Sequence analysis
Partial nucleotide (nt) sequence analysis of pMGA and pvpA genes from MG locals with reference strains were successfully accomplished using Bioedit and MEGA softwares and the summary of findings are shown in Table . Result from the analysis of sequence identity matrix and PCR product sizes of pMGA and pvpA genes are stated below.
Table 4. Analysis of pMGA and pvpA partial nucleotide sequences
3.4.1. pMGA gene
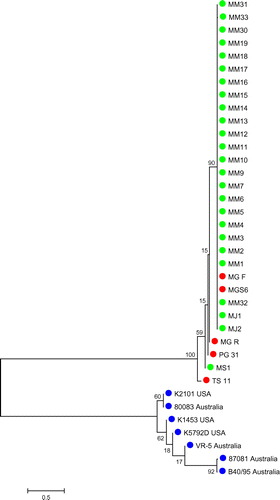
Upon the analysis of pMGA partial nucleotide sequences from field strains and five reference strains (MGS6, MG F, MG R, PG 31 and TS 11), results indicated that there were nucleotide variation and gene size polymorphism ranged between 226 and 336 bp having 29–34% of GC content. Although no nucleotide deletion was observed, nucleotide insertion of two field strains were found in the position of 139–156 nt. A total of 23 and 2 field strains showed 98–100% and 97–100% sequence similarity with MGS6 and TS 11 accordingly. Local field strains also showed some dissimilarity with strains of other countries regarding nucleotide variation. From the analysis of sequence identity matrix, the highest similarity was 0.987, which was observed in between MM14 and MM31 field strains. The lowest similarity was found in a field strain MS1 and an international strain 80,083 Australia. Overall, Malaysian local field strains showed maximum identity with the virulence vaccine strain MG F. Mention may be made that the similarity percentage of local field strains were very low to international strains indicated that Malaysian local strains were different from strains of other countries.
3.4.2. pvpA gene
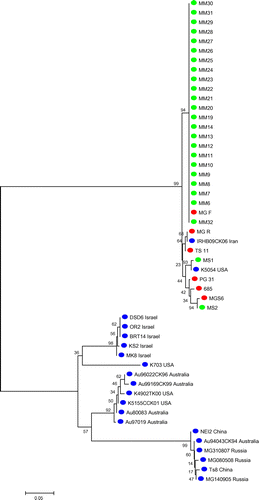
Upon the analysis of pvpA partial nucleotide sequence from field strains and six reference strains (MGS6, 6/85, MG F, MG R, PG 31 and TS 11), results indicated that there were nucleotide variation and gene size polymorphism ranged between 434 and 677 bp having 48–51% of GC content. There was a nucleotide insertion in one field strain in the position of from 1 to 65 nt and the nucleotide deletion was observed in the position of 573 nt to rest in 24 field strains. A total of 1, 14, 9 and 1 field strains showed 100%, 98–99, 99 and 98% sequence similarity with MGS6, TS 11, A5969 and PG 31, respectively. Local field strains also showed some dissimilarity with international strains regarding nucleotide variation. From the analysis of sequence identity matrix, the highest similarity value was 0.966, which was found in between field strains of MM19 with MM21 and MM14 with MM31. The lowest similarity value was 0.121, which was found in between field strains MM10 with MS2 and MM10 field strain with IRHB09CK06/Iran international strain. Mention also be made that the similarity value was very low between field strains and international strains, which indicated that Malaysian strains were different from strains of other countries.
3.5. Phylogenetic tree analysis
3.5.1. Phylogenetic analysis on the basis of pMGA partial nucleotide sequence
A phylogenetic tree was created on the basis of 226–336-bp partial nucleotide sequences of pMGA gene (Figure ). Upon the analysis of phylogenetic tree of pMGA partial nucleotide sequences from MG field, reference and international strains, results showed that a total of 24 out of 25 local strains were in the same cluster with the virulence vaccine strain MG F and the pathogenic reference strain MGS6. It also indicated that only the local strain MS1 was grouped with reference (MG R, PG 31 and TS 11) and international strains.
3.5.2. Phylogenetic analysis on the basis of pvpA partial nucleotide sequence
A phylogenetic tree was created on the basis of 434–677-bp partial nucleotide sequences of pvpA gene from local field, reference and international strains (Figure ). Upon the analysis of the phylogenetic tree of pvpA partial nucleotide sequences It had been illustrated that a total of 23 out of 25 Malaysian local strains were in the same group with the reference of high virulence vaccine strain MG F. were in different group from the pathogenic reference strain, MGS6 and less pathogenic vaccine strain, TS11 and located in the same cluster with two international strains. Only the two field strains (MS1 and MS2) placed in same group with international strains (K5054 USA and IRHB09CK06 Iran) and other reference strains (MG R, TS 11, MGS6, 6/85 and PG 31). It also showed that other international strains were grouped separately as there was no similarity found to Malaysian field strains.
4. Discussion
Mycoplasma gallisepticum (MG) is an important and most pathogenic avian Mycoplasma which is responsible for the great economic losses worldwide as it transmits vertically and horizontally. In this study, a total of 25 MG local field strains with reference strains for both of pMGA and pvpA genes were selected for gene targeted sequence analysis amplified by using published primers on the basis of pMGA and pvpA partial nucleotide sequence, respectively. The present study indicates the presence of pMGA and pvpA gene sequences and size variations among MG field strains from Malaysia. This size variation or gene size polymorphism is controlled by Mycoplasmas genetic mechanisms which depend on the combination of appropriate gene subsets encoding surface proteins having short homo- or heteropolymorphic tracts, undergoing intermittent and transformable changes in nucleotide numbers (Meseguer, Citation2008).
This finding concurred with other study where MG 6/85 strain was not amplified by AUTS11 F + ATTS11 R primer set of pMGA gene (Yasmin et al., Citation2014). Although these data were different from another study where one field strain from USA was amplified by AUAT TS 11 primer set showed similar gene size pattern with TS 11 and the vaccine strain 6/85 also amplified by this primer set but these two strains were different from each other due to the different gene size pattern (Kleven, Citation2002). The overall range of PCR product size was 434–677 bp when amplified by pvpA 1F + pvpA 2R primer set of pvpA gene which possessed some similarities with previous studies where pvpA gene showed size polymorphisms, with PCR products of 437, 578, 606 and 665 bp as detected among MG reference strains and local strains (Boguslavsky et al., Citation2000; Liu et al., Citation2001). Another researcher also showed the largest deletion found was that of a 230-bp fragment in vaccine strain F (Liu et al., Citation2001). Previous sequence analysis showed that MG reference strain R yield a 665-bp PCR product (Boguslavsky et al., Citation2000); which indicated similarity to the data found in this study where MG R reference strain yield 668-bp product size after amplification by pvpA 1F + pvpA 2R primer set of the gene pvpA. In this study, a total of 20 out of 25 local field strains showed 434–457-bp product size which is totally different from other study carried out in Malaysia, where it showed that, Malaysian local strains isolated from commercial chickens exhibited 567–611-bp PCR product when amplified by pvpA 1F + pvpA 2R primer set of pvpA gene (Zahraa et al., Citation2011). However, the findings of this study showed similarity with another study which stated that, Malaysian local strains exhibited gene size polymorphism at 437-bp PCR products (Kartini, Citation2012).
The partial nucleotide sequence analysis of pMGA and pvpA gene on MG local strains demonstrated that, there were nucleotide variation, size polymorphism, insertion and deletion, where pMGA showed similarities but pvpA gene showed dissimilarities with other study carried out in Malaysia, as in this study, in case of pvpA gene, most of the field strains showed 434–457-bp PCR product but Zahraa et al. (Citation2011) found 567–611 bp. Nucleotide sequence data also demonstrated that, there were insertion when amplified by pMGA partial nucleotide sequence primer and deletion when amplified by pvpA partial nucleotide sequence primer set which was similar to the previous studies (Kartini, Citation2012; Zahraa et al., Citation2011).
Findings of this study showed similarity with other study which stated that, MG-positive samples were closely related with MG F and MGS6 reference strain and distant from TS 11 vaccine strain (Zahraa et al., Citation2011). However, another study demonstrated that, MG local strains were distant from the reference strain (Kartini, Citation2012). There was no similarity found between Malaysian local strains and international strains (Figures and ). These findings are different from the other study which stated that, MG-positive field samples were distinct from vaccine reference strain (Zahraa et al., Citation2011). However, Kartini (Citation2012) showed that, MG-positive samples had similarity with MGS6 and had no similarity with MG F and MG R. Most of the international strains showed no similarities with Malaysian local strains.
5. Conclusion
Based on the phylogenetic tree analysis of pMGA and pvpA partial nucleotide sequences of MG local strains, reference strains and international strains, it can be concluded that Malaysian local strains are different from the international strains. This is an important information in relation to the types of vaccines to be used for the prevention of Mycoplasma infection. It would be necessary for further research on other different types of gene (IGSR) for characterization of MG field strains in Malaysia. A more efficacious vaccine may also be developed to control MG in Malaysia, based on the identified characteristics of MG field strains.
Funding
This work was financially supported by the Universiti Putra Malaysia and Ministry of Science, Technology and Innovation (MOSTI) [project number 02-01-04-SF0370].
Additional information
Notes on contributors
Farhana Yasmin
Farhana Yasmin is a dynamic, enthusiastic and hardworking veterinary professional who practices on ruminants and birds. She has done DVM and MS from the Hajee Mohammad Danesh Science and Technology University (HSTU), Bangladesh and Universiti Putra Malaysia (UPM), Malaysia respectively. In her master’s programme, she worked in the field of molecular biology and technically, she has gained skills on sample collection and preservation, culture, DNA extraction, DNA cloning, conventional PCR and sequencing with phylogenetic analysis and real-time PCR with efficiency and specificity for the thesis concerning “Development of a Diagnostic Real Time PCR Assay, Molecular Detection and Characterization of Mycoplasma gallisepticum”. This paper was culled from the MS thesis supervised by Aini Ideris, Abdul Rahman Omar and Mohd Hair-Bejo. Tan Sheau Wei, Tan Ching Giap, Rakibul Islam and Kartini Ahmad were her labmates and research colleagues who helped to collect the data and write the manuscript.
References
- Baseggio, N., Glew, M. D., Markham, P. F., Whithear, K. G., & Browning, G. F. (1996). Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology, 142, 1429–1435.10.1099/13500872-142-6-1429
- Bencina, D. (2002). Haemagglutinins of pathogenic avian mycoplasmas. Avian Pathology, 31, 535–547.10.1080/0307945021000024526
- Boguslavsky, S., Menaker, D., Lysnyansky, I., Liu, T., Levisohn, S., Rosengarten, R., … Yogev, D. (2000). Molecular characterization of the Mycoplasma gallisepticum pvpA gene which encodes a putative variable cytadhesin protein. Infection and Immunity, 68(7), 3956–3964.10.1128/IAI.68.7.3956-3964.2000
- Enright, M. C., & Spratt, B. G. (1999). Multilocus sequence typing. Trends in Microbiology, 7, 482–487.10.1016/S0966-842X(99)01609-1
- Ferguson, N. M., Hepp, D., Sun, S., Ikuta, N., Levisohn, S., Kleven, S. H., & Garcia, M. (2005). Use of molecular diversity of Mycoplasma gallisepticum by gene-targeted sequencing (GTS) and random amplified polymorphic DNA (RAPD) analysis for epidemiological studies. Microbiology, 151, 1883–1893.
- Ghorashi, S. A., Noormohammadi, A. H., & Markham, P. F. (2010). Differentiation of Mycoplasma gallisepticum strains using PCR and high-resolution melting curve analysis. Microbiology, 156(4), 1019–1029.
- Glew, M. D., Browning, G. F., Markham, P. F., & Walker, I. D. (2000). pMGA phenotypic variation in Mycoplasma gallisepticum occurs in vivo and is mediated by trinucleotide repeat length variation. Infection and Immunity, 68, 6027–6033.10.1128/IAI.68.10.6027-6033.2000
- Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper presented at the Nucleic Acids Symposium Series, 41, 95–98.
- Hosseini, H., Bozorgmehrifard, M. H., Peighambari, S. M., Pourbakhsh, S. A., & Razzazian, M. (2006). Random amplified polymorphic DNA (RAPD) fingerprinting of Mycoplasma gallisepticumisolates from chickens. Archive of Razi Institute, 61(2), 67–71.
- Jiang, H.-X., Chen, J.-R., Yan, H.-L., & Zeng, Z.-L. (2009). Molecular variability of DR-1 and DR-2 within the pvpA gene in Mycoplasma gallisepticum isolates. Avian Diseases, 53(1), 124–128.
- Kartini, A. (2012). Detection and molecular characterization of Mycoplasma gallisepticum and Mycoplasma synoviae from commercial chickens in Malaysia (pp. 59–112, Chapter 4. Master Thesis). Universiti Putra Malaysia, Serdang.
- Kleven, S. H. (2002). Recent developments in Mycoplasma diagnosis and control. Proceedings of the Western Poultry Disease Conference, 51, 109–113.
- Levisohn, S., Rosengarten, R., & Yogev, D. (1995). In vivo variation of Mycoplasma gallisepticum antigen expression in experimentally infected chickens. Veterinary Microbiology, 45, 219–231.10.1016/0378-1135(95)00039-D
- Liu, T., Garcia, M., Levisohn, S., Yogev, D., & Kleven, S. (2001). Molecular variability of the adhesin-encoding gene pvpA among Mycoplasma gallisepticum strains and its application in diagnosis. Journal of Clinical Microbiology, 39, 1882–1888.10.1128/JCM.39.5.1882-1888.2001
- Markham, P. F., Glew, M. D., Whithear, K. G., & Walker, I. D. (1993). Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infection and Immunity, 61, 903–909.
- Meseguer, M. A. (2008). Evolution of Mycoplasma pneumoniae and mycoplasmal infections. In F. Baquero, C. Nombela, & J. A. Gutierrez (Ed.), Evolutionary biology of bacterial and fungal pathogens (pp. 543–554). Washington, DC: ASM Press.
- Nascimento, E. R., Yamamoto, R., Herrick, K. R., & Tait, R. C. (1991). Polymerase chain reaction for detection of Mycoplasma gallisepticum. Avian Diseases, 35, 62–69.10.2307/1591296
- Papazisi, L., Gorton, T. S., Kutish, G., Markham, P. F., Browning, G. F., Swartzell, S., … Geary, S. J. (2003). The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain rlow. Microbiology, 149, 2307–2316.10.1099/mic.0.26427-0
- Razin, S., Yogev, D., & Naot, Y. (1998). Molecular biology and pathogenicity of mycoplasmas. Microbiology and Molecular Biology Reviews, 62, 1094–1156.
- Sasaki, Y., Ishikawa, J., Yamashita, A., Oshima, K., Kenri, T., Furuya, K., … Sasaki, T. (2002). The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Research, 30, 5293–5300.10.1093/nar/gkf667
- Sprygin, A. V., Elatkin, N. P., Kolotilov, A. N., Volkov, M. S., Sorokina, M. I., Borisova, A. V., … Drygin, V. V. (2011). Biological characterization of Russian Mycoplasma gallisepticum field isolates. Avian Pathology, 40(2), 213–219.
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.10.1093/molbev/msr121
- Tan, C. G. (2008). Gene size polymorphism and pathogenicity in embryonated eggs of Mycoplasma gallisepticum isolated from commercial chickens (pp. 31–165, Chapters 3, 4 and 5. Master Thesis). Universiti Putra Malaysia, Serdang.
- Thompson, J. D., Higgins, D. G., Gibson, T. J., & Clustal, T. J. (1994). Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.10.1093/nar/22.22.4673
- Yasmin, F., Ideris, A., Omar, A. R., Hair-Bejo, M., Tan, S. W., Tan, C. G., … Ahmad, K. (2014). Molecular detection of Mycoplasma gallisepticum by real time PCR. Jurnal Veterinar Malaysia, 26(1), 1–7.
- Zahraa, F., Aini, I., Omar, A. R., Hair-Bejo, M., & Tan, C. G. (2011). Molecular identification of two genetic markers that distinguish between pathogenic and nonpathogenic strains of Mycoplasma gallisepticum. Animals and Veterinary Advances Journal, 10, 2846–2855.