ABSTRACT
It is clear that several prostate cancers remain indolent whereas others develop into advanced forms. There is a need to improve patient management by identifying biomarkers for personalized treatment. We demonstrated that miR-15/miR-16 loss, miR-21 upregulation, and deregulation of their target genes represent a promising predictive signature of poor patient prognosis.
KEYWORDS:
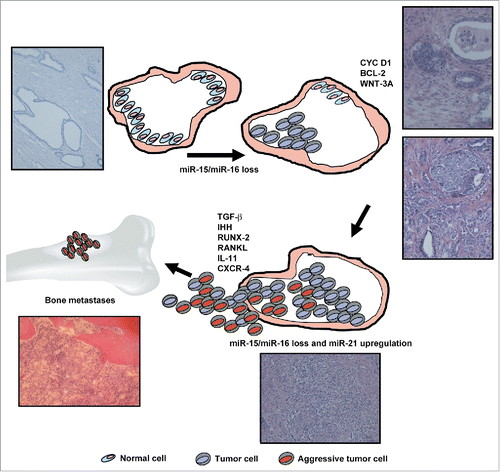
The era of high-resolution whole genome and transcriptome sequencing technologies has revealed at least 90% of the genome is actively transcribed in non-coding RNAs whereas protein coding genes represent less than 2% of total sequences.Citation1 Non-coding RNAs are extremely stable in tissues and biological fluids and are emerging as new causative players in diseases, including cancers. Because of their abundance and stability, they represent a new source for discovering novel therapeutic approaches and biomarkers for improving tumor diagnosis and patient management. Non-coding RNAs may be roughly grouped into 2 major classes based on transcript size: small (18–200 nt) and long (200 nt to > 100 kb) RNAs.Citation1 Small RNAs include the well-documented microRNA (miRNA or miR) gene family. These molecules are involved in the specific regulation of both protein-coding and non-coding genes by transcriptional and post-transcriptional silencing; in particular, several lines of evidence show that they are sophisticated regulators of oncogenes and tumor suppressor genes. Prostate cancer is the most frequent tumor in men; however, over the last decade clinical reports have shown that a high percentage of these tumors remain indolent while a constant fraction progress to advanced therapy-resistant forms. During the last few years the prostate specific antigen (PSA) test has improved tumor diagnosis but it has also resulted in overtreatment and has not reduced patient death. Mutations, amplifications, deletions and fusions of genes such as epidermal growth factor receptor (EGFR), phosphatase and tensin homolog (PTEN), V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (K-RAS), anaplastic lymphoma kinase (ALK), mesenchymal-epithelial transition factor (MET), BCR-ABL, progressive multifocal leukoencephalopathy (PML), and retinoic acid receptor α (RARα) are conventionally used to drive the therapeutic approach for several types of cancer.Citation2 In stark contrast, the management of prostate cancer patients still lacks molecular indicators. Therefore, this field can hugely benefit from the discovery of biomarkers. Several years ago, we demonstrated that the cluster miR-15a/miR-16 is a tumor suppressor in prostate cancer.Citation3 This cluster resides at the 13q14.3 genomic region that is frequently deleted in high-stage tumors. We hypothesized that during cancer progression the cluster may be aberrantly silenced or deleted and demonstrated that its downregulation per se caused cancer development and progression. In the following years, many articles have highlighted the tumor suppressing role of these 2 miRNAs in different types of cancer.Citation4 In the paper by Bonci et al. (Oncogene June 2015), we investigated the effect of miR-15 and miR-16 loss on prostate cancer metastatic spread. We forced miR-15 and miR-16 downregulation in RWPE-2 cells, a cell line representative of early tumors that is unable to form metastases in murine models. Surprisingly, these tumor cells acquired a metastatic phenotype, selectively invading bones. Since RWPE-2 cells were transformed by K-RAS, we induced that loss of miR-15/miR-16 might synergize with RAS activation to promote bone colonization with subsequent osteosclerosis and osteolysis thus recapitulating the bone lesions of human patients in immunocompromised mice. miR-21 overexpression resembles RAS aberrant activity,Citation5 and we demonstrated that RAS increases miR-21 expression in prostate cancer cells. Non-transformed prostate cells engineered to express high levels of miR-21 and low levels of miR-15/miR-16 acquired invasion properties and produced bone metastases, demonstrating that such aberrant miRNA expression per se is sufficient to induce an aggressive phenotype. We observed a consistent increase of miR-21 in primary cells isolated from patients with loss of miR-15/miR-16. Such concomitant alterations of miRNA levels may have detrimental effects in patients who do not undergo rapid tumor debulking, as suggested by the striking correlation between this miRNA pattern and prostate cancer progression in in silico analysis of a gene set array by Taylor et al.Citation6
The cooperation between increased miR-21 and loss of miR-15/miR-16 seems to particularly occur at the level of transforming growth factor β (TGF-β) signaling. Notably, from in silico analysis several targets of miR-15, miR-16, and miR-21 are related to the TGF-β pathway. miR-15 and miR-16 can target ACTIVIN RIIA, morphogens belonging to the same family that is triggered by ACTIVIN A and NODAL. The increased expression of NODAL reported in prostate cancer may therefore contribute to enhanced SMAD signaling after loss of miR-15 and miR-16. In addition, miR-21 controls SMAD-7, an inhibitor of the TGF-β pathway. We present a new molecular circuit that is driven by alterations in miR-15, miR-16, and miR-21 and results in aberrant TGF-β signaling (). Several bone metastasis-associated genes induced by TGF-β are indirectly affected, such as receptor activator of nuclear factor kappa-B ligand (RANKL), runt-related transcription factor 2 (RUNX2), C-X-C chemokine receptor type 4 (CXCR-4), connective tissue growth factor (CTGF), and interleukin 11 (IL-11). It has been reported that TGF-β can post-transcriptionally regulate indian hedgehog (IHH) ligand,Citation7 a key gene in bone metastasis formation.Citation8 We demonstrated that miR-15 and miR-16 can directly control the IHH gene. The results showed an aberrant pro-metastasis circuit involving TGF-β, IHH, and miRNA alterations. Although considerable efforts have been made to identify patients at high risk of recurrence, currently available risk stratification models and predictive nomograms lack adequate accuracy. The proposed changes in miRNA levels correlate with poor prognosis but they do not significantly correlate with higher Gleason scores or PSA levels, suggesting that this signature may add further information to conventional parameters. Administration of targeted or conventional therapies requires accuracy of staging procedures and biomarkers predictive of patient response.Citation9 miR-15/miR16 and miR-21 control several gene pathways that are key targets of FDA-approved drugs (such as denosumab or TGF-β and IHH inhibitors). A good candidate biomarker should be functionally correlated with pathology, retain several properties such as stability and reliability, and be analyzable with a non-invasive, reportable, and easy-to-handle technical approach. To address all these requirements a signature of multiple elements seems to be more appropriate. Much evidence shows that tissue- or blood-based miRNA biomarkers that predict clinical behavior and/or therapeutic response can be used as prognostic and predictive indicators. Moreover, targeting of disseminated tumorigenic cells before formation of the protective metastatic niche appears to be a promising novel therapeutic strategy in cancer.Citation10 For the above reasons, our data suggest that miR-15 and miR-16 downregulation combined with miR-21 upregulation should be investigated further to verify whether these molecular parameters increase the accuracy of current predictors. Furthermore, the multiple molecular abnormalities related to deregulation of these RNA molecules suggest a role as predictive biomarkers for optimal testing of innovative molecular targeted agents and bone-acting compounds in patients. Our data may offer a rationale for clinical trials of biomarker-based prevention of bone metastasis. This information suggests a new molecular signature for optimizing the management of prostate cancer and may implicate new druggable pathways for the treatment of bone metastases.
Figure 1. Molecular mechanisms whereby alterations in miR-15, miR-16, and miR-21 result in aberrant TGF-β signaling. Hematoxylin and Eosin staining of patient tissues is reported. BCL-2, B-cell lymphoma; CXCR-4, C-X-C chemokine receptor type 4; CYC D1, cyclin D1; IHH, Indian hedgehog; IL-11, Interleukin 11; RUNX-2, Runt-related transcription factor 2; RANKL, Receptor activator of nuclear factor kappa-B ligand; TGF-β, Transforming growth factor β; WNT-3A, Wingless-Type MMTV Integration Site Family, Member 3A.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Giuseppe Loreto and Tania Merlino for their technical support.
Funding
This manuscript has been supported by National Ministry of Health, Under-40 researcher, and Italy-USA microRNA program to DB, and the Italian Association for Cancer (AIRC) and Fondazione Roma funding to RDM.
References
- Lieberman J, Slack F, Pandolfi PP, Chinnaiyan A, Agami R, Mendell JT. Noncoding RNAs and cancer. Cell; 153:9-10; PMID:23781554
- Sawyers C. Targeted cancer therapy. Nature 2004; 432:294-7; PMID:15549090; http://dx.doi.org/10.1038/nature03095.
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 2008; 14:1271-7; PMID:18931683; http://dx.doi.org/10.1038/nm.1880
- Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ; 17:215-20; PMID:19498445; http://dx.doi.org/10.1038/cdd.2009.69
- Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell; 18:282-93; PMID:20832755; http://dx.doi.org/10.1016/j.ccr.2010.08.013
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell; 18:11-22; PMID:20579941; http://dx.doi.org/10.1016/j.ccr.2010.05.026
- Murakami S, Nifuji A, Noda M. Expression of Indian hedgehog in osteoblasts and its posttranscriptional regulation by transforming growth factor-β. Endocrinology 1997; 138:1972-8; PMID:9112395; http://dx.doi.org/10.1210/endo.138.5.5140
- Sanchez P, Clement V, Ruiz i Altaba A. Therapeutic targeting of the Hedgehog-GLI pathway in prostate cancer. Cancer Res 2005; 65:2990-2; PMID:15833820; http://dx.doi.org/10.1158/0008-5472.CAN-05-0439
- Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: emerging therapeutic strategies. Nat Rev Clin Oncol 2011; 8:357-68; PMID:21556025
- Zeuner A, De Maria R. Not so lonely at the top for cancer stem cells. Cell Stem Cell 2011; 9:289-90; PMID:21982226; http://dx.doi.org/10.1016/j.stem.2011.09.006
