ABSTRACT
DNA termini at double-strand breaks are often chemically heterogeneous and require processing before initiation of repair. In a recent report, we demonstrated that CtIP and the MRE11-RAD50-NBS1 (MRN) nuclease complex cooperate with BRCA1 to specifically repair topoisomerase II-DNA adducted breaks. In contrast, BRCA1 is dispensable for repair of restriction endonuclease-generated double-strand breaks.
Defective DNA repair results in infertility, immunological defects, cancer, and neurodegenerative syndromes. In particular, DNA double-strand breaks (DSBs) can lead to loss of genetic material or chromosomal rearrangements if not repaired or misrepaired. This leads to genomic instability, which in turn favors tumorigenesis. Indeed, many tumors display alterations (overexpression, loss of function, or mutations) in proteins involved in DSB repair.Citation1-3
DSBs can arise from endogenous sources, in particular DNA transactions such as DNA replication, DNA recombination, and RNA transcription, as well as clastogens including ionizing radiation and DNA-damaging chemicals. DNA termini at DSBs are often chemically heterogeneous and thus require processing before the final ligation step common to all DSB repair pathways—non-homologous end-joining (NHEJ) or homology-directed repair (HDR)—can restore DNA integrity. Persistent DSBs that are difficult to repair are a particular threat to genome integrity as they are more likely to interfere with DNA replication forks or elongating RNA polymerases and to facilitate aberrant recombinogenic events. Whereas ligatable “clean” breaks are rapidly and efficiently repaired by NHEJ, chemically heterogeneous breaks such as protein-DNA adducts require dedicated enzymes for multistep processing.
DNA topoisomerases are enzymes that resolve DNA topological stress arising during DNA replication, transcription, recombination, and repair. Type IIA topoisomerases (Top2) transiently generate a DSB to allow passage of a second DNA duplex through the break. Incomplete topoisomerase reactions that stabilize the transient protein–DNA adduct are a significant source of DSBs in unperturbed cells.Citation4 Etoposide is a topoisomerase poison that binds to the topoisomerase-DNA adduct and increases its half-life by inhibiting DNA religation. This in turn blocks DNA transactions including the initiation of DNA end resection, the step that initiates HDR. As cancer cells display elevated replication rates and rely heavily on DNA repair pathways, topoisomerase poisons are widely used as chemotherapy drugs to treat many tumors although resistance typically eventually develops.Citation1
Eukaryotic cells use two pathways to process and repair Top2-DNA crosslinks. In the first pathway, ubiquitin-dependent degradation of Top2 is coupled with a tyrosyl-DNA phosphodiesterase (TDP2, also known as TTRAP) to remove the residual phosphotyrosine linkages. A second pathway, first uncovered from genetic studies in yeast, implicates the MRX/MRN complex (Mre11-Rad50-Xrs2 in yeast; MRE11-RAD50-NBS1 in higher eukaryotes) and Sae2 protein (RBBP8 in humans, more commonly known as CtIP) in the nucleolytic release of 5′-linked proteins; this includes removal of the topoisomerase-related Spo11, which forms stable protein-DNA adducts to initiate meiotic recombination.Citation1,5,6
In our recent publication “MRN, CtIP, and BRCA1 mediated repair of topoisomerase II–DNA adducts”,Citation7 we delineated the contribution of the second pathway to the removal of Top2-DNA covalent adducts generated by the chemotherapeutic drug etoposide and their subsequent repair in a cell-free system derived from Xenopus egg extracts.
Genetic studies in yeast suggested a role of the MRX/N complex and Sae2/CtIP in tolerance to damage caused by topoisomerase poisons.Citation5,8 Studies in chicken and mammalian tissue culture cells also showed that loss of MRN, CtIP, or the tumor suppressor breast cancer 1 (BRCA1) enhanced sensitivity to topoisomerase poisons. To complement these studies, we developed a unique set of biochemical approaches to address the mechanistic basis of Top2-DNA adduct removal and processing and specifically the role of BRCA1, which is absent in yeast. Xenopus cell-free extracts allow for the complete inactivation of essential proteins such as MRE11, CtIP, or BRCA1.
We developed the following biochemical assays and readouts:
We monitored chromosomal replication in extracts treated with a small dose of etoposide that does not affect DNA replication in control undepleted extracts.
We directly monitored the removal of Top2-DNA adducts by isolating genomic DNA following cesium chloride gradient centrifugation and probing against topoisomerase 2.
We assessed resection, the first step of HDR, by monitoring single-stranded DNA–Replication Protein A (ssDNA-RPA) intermediates following Top2 inhibition.
We found that removal of the MRN complex (by depleting the MRE11 subunit), depletion of CtIP, or depletion of BRCA1 similarly (1) strongly inhibited genomic DNA replication in the presence of a low dose of etoposide; (2) resulted in the persistence of Top2-DNA adducts in the presence of etoposide; and (3) significantly inhibited DNA resection at etoposide-induced DSBs. This indicated that the MRN-CtIP-BRCA1 pathway is required for tolerance to etoposide during DNA replication, for Top2-DNA adduct removal, and for subsequent processing of DNA ends to generate a 3´ ssDNA overhang and further suggested that Top2-DNA adduct removal by MRN-CtIP is analogous to the nucleolytic removal of SPO11 covalent complexes during meiotic recombination. In future studies, we will attempt to isolate Top2-oligonucleotide complexes from etoposide-treated genomic DNA, similar to the previously described SPO11-bound oligonucleotides.Citation6
Notably, we uncovered specific requirements for processing etoposide-induced breaks compared to endonuclease-generated breaks.
In contrast to depletion of MRN, CtIP, or BRCA1, all of which impaired processing of Top2-DNA adducts, depletion of exonuclease 1 (EXO1) did not impact Top2 removal. This establishes that EXO1 does not participate in Top2 removal.
We used a mutant form of CtIP (S328A) that cannot interact with BRCA1 yet supports resection of endonuclease-generated breaks in CtIP-depleted extracts. This mutant does not promote Top2 removal and resection from Top2-induced DSBs, demonstrating that the CtIP–BRCA1 interaction is specifically required for MRN-CtIP–mediated removal of Top2-DNA adducts and initiation of resection.
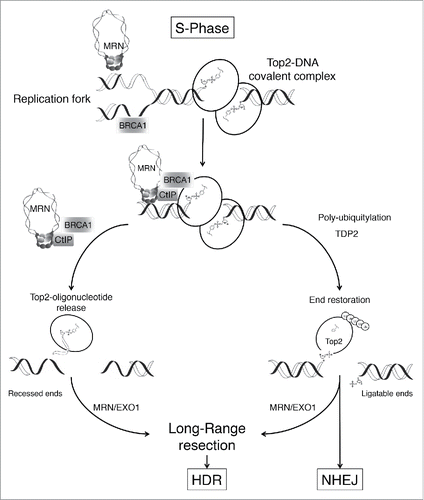
Together, our biochemical analyses explain the specific hypersensitivity of cells deficient in MRN, CtIP, or BRCA1 to etoposide poisons. Our work supports a role for the MRN-CtIP-BRCA1 pathway as a cell cycle-regulated nuclease activity that releases blocks on the DNA during replication and favors HDR (see ). We speculate that BRCA1 might enhance adduct recognition or help stabilize replication forks upon encountering adducts. Interestingly, we also found that BRCA1 was specifically recruited to Top2-DNA adduct-containing chromatin during replication. However, the exact mechanism by which CtIP-BRCA1 interaction facilitates the removal of Top2-DNA adducts warrants further investigation.
Figure 1. Putative mechanism for removal of Top2-DNA adducts. Nucleolytic processing by the MRE11-RAD50-NBS1 complex (MRN), CtIP, and BRCA1 represents a fast and efficient mechanism of type IIA topoisomerase (Top2)-DNA adduct removal that prevents catastrophic collisions with replication forks. The MRN-CtIP-BRCA1 pathway promotes subsequent double-strand break repair through homology-directed repair (HDR) by creating a substrate suitable for DNA end resection. We propose that during S-phase the MRN-CtIP-BRCA1 pathway favors HDR, the preferred double-strand break repair pathway during DNA replication. Outside S-phase, resection is less efficient and TDP2 promotes end restoration requiring Top2 polyubiquitination, Top2 denaturation, and/or proteasome-mediated degradation. However, this nucleolytic pathway can also operate in the absence of DNA replication. EXO1, exonuclease 1; NHEJ, non-homologous end-joining; TDP2, tyrosyl-DNA phosphodiesterase 2.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all members of the laboratory for useful discussions and apologize to authors whose works are not cited due to space restrictions.
Funding
This work was supported in part by National Cancer Institute grants CA092245, CA167826, and CA174653 to J. Gautier.
References
- Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ Mol Mutagen 2015; 56:1-21; PMID:25111769; http://dx.doi.org/10.1002/em.21892
- Aparicio T, Baer R, Gautier J. DNA double-strand break repair pathway choice and cancer. DNA Repair 2014; 19:169-75; PMID:24746645; http://dx.doi.org/10.1016/j.dnarep.2014.03.014
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet 2011; 45:247-71; PMID:21910633; http://dx.doi.org/10.1146/annurev-genet-110410-132435
- Liu LF, Rowe TC, Yang L, Tewey KM, Chen GL. Cleavage of DNA by mammalian DNA topoisomerase II. J Biol Chem 1983; 258:15365-70; PMID:6317692
- Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell 2009; 33:117-23; PMID:19150433; http://dx.doi.org/10.1016/j.molcel.2008.11.021
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nat Cell Biol 2005; 436:1053-7; PMID:16107854; http://dx.doi.org/10.1038/nature03872
- Aparicio T, Baer R, Gottesman M, Gautier J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II–DNA adducts. J Cell Biol 2016; 212:399-408; PMID:26880199; http://dx.doi.org/10.1083/jcb.201504005
- Malik M, Nitiss JL. DNA repair functions that control sensitivity to topoisomerase-targeting drugs. Eukaryot Cell 2004; 3:82-90; PMID:14871939; http://dx.doi.org/10.1128/EC.3.1.82-90.2004
