ABSTRACT
Network approaches are ubiquitous, from social and ecological systems up to complex biological processes. In our recently published work we used the network framework for a Systems Medicine approach to multiple cancer types, in order to highlight similitudes and differences that can be exploited to extend existing therapeutical strategies. These approaches shed new light to oncological processes, but allow also to pose “old” questions (like the search for novel drug targets) in a “new” way.
Networks and cancer
In our work “Network integration of multi-tumour omics data suggests novel targeting strategies”Citation1 we have integrated transcriptional (“phenotype”) and mutational (“genotype”) information from 11 different types of cancer from “The Cancer Genome Atlas” (TCGA) databaseCitation2 (Colon and Rectum Adenocarcinomas; Lung Adenocarcinoma, Lung Squamous Cell Carcinoma, Glioblastoma Multiforme, Ovarian Serous Cystadenocarcinoma, Breast Invasive Carcinoma, Uterine Corpus Endometrial Carcinoma, Brain Lower Grade Glioma, Kidney Renal Clear Cell and Papillary Cell Carcinomas) trying to grasp a wider picture of the mechanisms involved in the pathology beyond a single-tumour approach.
We used network methods throughout all our analysis pipeline: to cluster tumours based on functional relationship between genes (i.e. generating a “network” of transcriptional profile correlations for each tumour) and not based on patients’ similarity; to embed phenotypical and genotypical information in a Protein Interaction Network (BioplexCitation3-OntocancroCitation4 network), by combining high-throughput protein binding experimentsCitation3 and biological annotation of known cancer-related processes;Citation4 prioritizing drug targets based on their role in the constructed networks, by identifying the most central nodes (corresponding to genes) in our network.Citation5
Three tumour clusters were identified, reflecting cancer anatomical proximity (as for kidney and intestine carcinomas) but with some interesting difference (e.g. brain tumours not clustering together). From each cluster, a different network was constructed, starting from the same initial Bioplex-Ontocancro “template” network, but with different links depending on cluster-specific gene-gene coexpression levels (see ). From each network, a different set of central genes emerged: these signatures were derived from transcriptomics profiles, but were found in the proximity of known mutated genes, thus evidencing the “interest” of cancer in taking control of the underlying biological mechanisms (through mutations) to modify their functionality (via transcriptional changes). The expression profile of these signatures also allowed to stratify patients inside each cluster (thus independently on their tumour type) in terms of survival, showing a clear clinical relevance. Most signature genes were already known as drug targets, or were identified as objects of ongoing clinical trials. A comparative analysis of drugs targeting our signature genes, against drugs targeting genes in the same networks but not in the signature, was performed on more than 300 cell lines from tumours associated to our 11 tumour types:Citation6 in many cases (>62% in total, >95% in two of the three clusters) the IC50 (i.e. the concentration of a drug that gives half-maximal response) was lower for our selected targets. Also a small set of experiments was performed in vitro on two cell lines (MCF-7, a breast cancer cell line, and T98G, a glioblastoma cell line) showing better response only for signature drug targets.
Figure 1. Network of Cluster 2 signature genes (dark blue) and their first neighbors in the BioplexOntocancro network (yellow). In the same picture, known mutated genes (light blue) and known drug targets (light green) are shown. The red circles highlight six possible gene modules inside the network, that could reflect possible synergistic effects for drug targeting or deeper-level biological relationships.
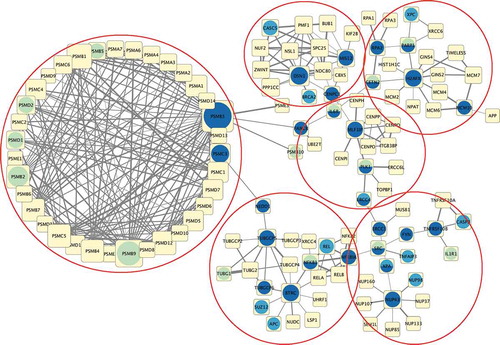
This analysis confirmed the validity of our approach for grouping tumours based on a more functional and structural (as schematized by the network framework) rather than a merely anatomical point of view, and allowed the identification of targets common to all tumours of the same cluster, that could in principle facilitate drug repurposing between these different tumour types.
We obtained some answer, but many more questions can stem from these analyses. We propose some of them in the final part of this commentary, with the idea that more information can be extracted from the constructed networks, and that further knowledge can be achieved by the addition of other types of data to the model. These studies will necessarily require highly interdisciplinary efforts, involving clinicians, biologists, statisticians, bioinformaticians and mathematical modeling (as provided, for example, by network theory):
Some of the signature genes had already been identified as drug targets, and thus could be directly tested for their efficacy against tumours. What about the other signature genes which are not actual drug targets?
We observed a synergistical effect between drugs acting on two signature genes: is synergy favored between neighboring genes in the Bioplex-Ontocancro network? (similar questions are starting to be recently posedCitation7)
What is the role of epigenetic and posttranscriptional regulation? We remark that DNA methylation and miRNA expression are available for most tumours in TCGA, and could thus be integrated into the model, or in the same network or as a further layer, in a multiplex network fashionCitation8,Citation9
Can we expand our results towards Precision medicine, based on individual mutational and transcriptional profile, in order to design individualized therapies with specific drugs taken singularly or in combination?
There is still a long way to a reliable in silico modeling of cancer(s), able to effectively impact clinical and pharmacological practice, but undoubtedly the Big Data Era is showing its effects also in the biomedical field: the increase in data availability, both in quantity and in data-type variety, looks promising, at least in prioritizing the list of possible trials to be pursued with full experimental depth.
Acknowledgments
D.R. acknowledges EU IMI-2 “HARMONY” project n. 116026, and “Interomics” CNR Flagship Initiative.
Additional information
Funding
References
- Do Valle I, Menichetti G, Simonetti G, Bruno S, Zironi I, Durso DF, Mombach JCM, Martinelli G, Castellani G, Remondini D. Network integration of multi-tumour omics data suggests novel targeting strategies. Nat Commun. 2018;9:4514. doi:10.1038/s41467-018-06992-7.
- The Cancer Genome Atlas. [accessed 2018 Dec]. https://cancergenome.nih.gov/.
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The bioplex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi:10.1016/j.cell.2015.06.043.
- Ontocancro. [accessed 2018 Dec]. http://ontocancro.inf.ufsm.br/.
- Pauls SD, Remondini D. Measures of centrality based on the spectrum of the Laplacian. Phys Rev E. 2012;85:066127. doi:10.1103/PhysRevE.85.066127.
- Genomics of Drug Sensitivity in Cancer. [accessed 2018 Dec]. http://www.cancerrxgene.org.
- Li H, Li T, Quang D, Guan Y. Network propagation predicts drug synergy in cancers. Cancer Res. 2018;78:18.
- Kivela M, Arenas A, Barthelemy M, Gleeson JP, Moreno Y. Multilayer networks. J Complex Networks. 2014;2:203–271.
- Menichetti G, Remondini D, Panzarasa P, Mondragon RJ, Bianconi G. Weighted multiplex networks. PLoS One. 2014;9(6):e97857. doi:10.1371/journal.pone.0097857.
