ABSTRACT
In budding yeast, Atg1 kinase, together with Atg13 and Atg17, forms a complex that is essential for autophagy. Previous work showed that the Atg1 kinase complex is regulated extensively by phosphorylations. Our recent paper demonstrates that type 2C protein phosphatases Ptc2 and Ptc3 are involved in the dephosphorylation of Atg13 and Atg1 kinase to promote autophagy.
KEYWORDS:
Macroautophagy (hereafter referred to as autophagy) is a catabolic pathway which is involved in vacuolar degradation of random portions of the cytoplasm in response to nutrient starvation.Citation1 In budding yeast, the Atg1–Atg13–Atg17 kinase complex is essential for autophagy. Under basal conditions, the target of rapamycin complex 1 (TORC1), a sensor of nutrient availability, is active and maintains Atg13 in an inactive and hyperphosphorylated state.Citation2 Upon the nutrient starvation, TORC1 activity is inhibited, and Atg13 is rapidly dephosphorylated. The dephosphorylation of Atg13 is necessary for the induction of autophagy because expression of a constitutively active ATG13-8SA allele leads to autophagy induction uncoupled from TORC1 activity.Citation2 As with Atg13, Atg1 is shown to be regulated extensively by phosphorylations.Citation3 However, the phosphatases that are involved in the reversal of these phosphorylations remained elusive.
In addition to the targeting of random cargo to the vacuoles through starvation-induced macroautophagy, distinct cargo can be targeted for degradation under nutrient-replete conditions through selective autophagy. Selective autophagy relies on the binding of cargo to the specific autophagy receptors and then to the adapter protein Atg11.Citation4 Recent work showed that the interactions between Atg11-bound cargo can stimulate Atg1 kinase activity and lead to Atg1 activation.Citation5
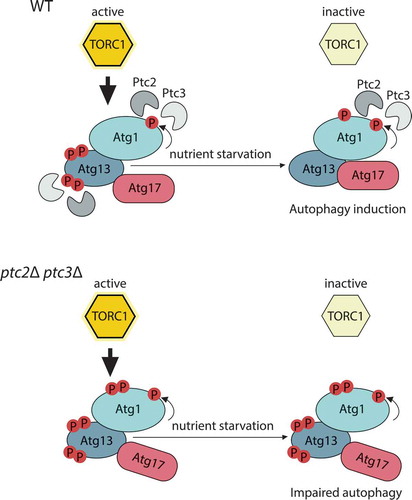
In our recent study,Citation6 we characterized two redundant type 2C protein phosphatases (PP2C), Ptc2 and Ptc3, as novel regulators of Atg1 kinase complex (). Deleting both of these phosphatases leads to impaired autophagy function and impaired delivery of the autophagy cargo to vacuoles. This defect is not specific to starvation-induced macroautophagy, as the ptc2Δ ptc3Δ double mutant is also defective in at least two selective autophagy pathways: the cytoplasm-to-vacuole targeting (Cvt) pathwayCitation7 and genotoxin-induced targeted autophagy.Citation8 The essential Atg machinery is mislocalized in the absence of Ptc2 and Ptc3, which could be the reason for the selective autophagy defect.
Figure 1. Ptc2 and Ptc3 promote autophagy through dephosphorylation of Atg1 kinase complex.
In wild-type (WT) cells, under nutrient-deplete conditions, target of rapamycin complex 1 (TORC1) is active, and Atg13 is kept in a hyperphosphorylated and inhibited state (upper panel). Atg1 kinase activity is stimulated by Atg11-bound cargo interactions, which lead to Atg1 phosphorylation. Ptc2 and Ptc3 counteract these modifications. Upon TORC1 inhibition by nutrient starvation, Ptc2 and Ptc3 contribute to the dephosphorylation of Atg13, and Atg1–Atg13–Atg17 complex formation, to allow proper autophagy induction. The deletion of these phosphatases (lower panel) leads to the accumulation of hyperphosphorylated Atg1 and Atg13 species and impaired Atg13–Atg17 interaction.
Next, we sought to identify the targets of Ptc2 and Ptc3 phosphatases among the Atg machinery to understand the mechanism of autophagy regulation by these phosphatases. We found that TORC1 function is not impaired in ptc2Δ ptc3Δ mutant, as measured by Sch9 phosphorylationCitation;9 therefore, we focused on the Atg proteins downstream of TORC1. Similar to what we observe under the basal conditions, the essential Atg machinery is also mislocalized in the ptc2Δ ptc3Δ mutant following TORC1 inhibition. Additionally, deletion of Ptc2 and Ptc3 results in an accumulation of higher molecular weight Atg13 bands. Overexpression of an ATG13-8SA allele from a plasmid fully rescues the autophagy defect of ptc2Δ ptc3Δ mutant; moreover, expression of ATG13-8SA as a single copy from endogenous ATG13 locus also partially rescues the autophagy defect. Based on these data, we conclude that Ptc2 and Ptc3 phosphatases promote autophagy, at least in part, through dephosphorylation of Atg13.
In the ptc2Δ ptc3Δ double mutant, we also detect an accumulation of slower-migrating Atg1 bands under basal conditions, which persists after nutrient depletion. The appearance of these bands is dependent on the Atg13 and Atg11 and the kinase activity of Atg1. Therefore, we conclude that these slower-migrating Atg1 species are a result of increased Atg1 autophosphorylation in the absence of Ptc2 and Ptc3. However, the accumulation of autophosphorylated Atg1 does not inhibit autophagy, as overexpression of constitutively active ATG13-8SA allele in ptc2Δ ptc3Δ mutant did not abolish the appearance of these hyperphosphorylated Atg1 bands, even though it rescued the autophagy defect. Surprisingly, the accumulation of higher molecular weight Atg1 and Atg13 species in the absence of Ptc2 and Ptc3 did not impair the Atg1–Atg13 interaction. However, the Atg13–Atg17 interaction in ptc2Δ ptc3Δ was approximately 50% reduced compared to a wild-type control.
We observe that immunoprecipitating Ptc2 and Ptc3 enriched for Atg1, Atg13, and Atg17, suggesting that these two phosphatases interact with the Atg1 kinase complex. The interaction of the PPC2 phosphatases with the Atg1 complex is detectable even under nutrient-replete conditions. This is in agreement with our findings demonstrating the accumulation of slower-migrating Atg1 and Atg13 species under nutrient-replete conditions, and it implies that the regulation of Atg1 kinase complex by these two phosphatases is constitutive. Interestingly, after autophagy induction, we detect a reduction in Ptc2 and Ptc3 protein abundance. Furthermore, Ptc2 and Ptc3 are rapidly depleted after inhibition of protein synthesis, suggesting that they are short-lived. We hypothesize that the degradation of these phosphatases could be one way of maintaining hyperphosphorylated Atg13 rapidly once TORC1 is activated.
Ptc2 and Ptc3 have been studied extensively in the context of DNA damage and stress response.Citation10 In our recent study, we find that they are also critical for the regulation of autophagy. Taking previously published work into account,Citation5 these findings suggest that Atg1 kinase activity is stimulated by the receptor-bound cargo interactions constitutively, and phosphatases Ptc2 and Ptc3 counteract these phosphorylations. Atg13 is also hyperphosphorylated in the absence of Ptc2 and Ptc3, which could be the underlying reason for the impaired Atg13–Atg17 interaction. Because the expression of a genomic single copy ATG13-8SA allele only partially rescues the autophagy defect of ptc2Δ ptc3Δ mutant, we conclude that other residues on Atg13 or other Atg proteins such as Atg17 could be subject to regulation by Ptc2 and Ptc3.
Additional information
Funding
References
- Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016;428(9 Pt A):1681–1699. doi:10.1016/j.jmb.2016.02.021.
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058. doi:10.1128/MCB.01344-09.
- Yeh YY, Shah KH, Chou CC, Hsiao HH, Wrasman KM, Stephan JS, Stamatakos D, Khoo KH, Herman PK. The identification and analysis of phosphorylation sites on the Atg1 protein kinase. Autophagy. 2011;7:716–726.
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16(4):1593–1605. doi:10.1091/mbc.e04-11-1035.
- Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell. 2015;59(3):372–381. doi:10.1016/j.molcel.2015.06.009.
- Memisoglu G, Eapen VV, Yang Y, Klionsky DJ, Haber JE. PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Proc Natl Acad Sci U S A. 2019;116(5):1613–1620. doi:10.1073/pnas.1817078116.
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS letters. 2010;584(7):1359–1366. doi:10.1016/j.febslet.2010.02.013.
- Eapen VV, Waterman DP, Bernard A, Schiffmann N, Sayas E, Kamber R, Lemos B, Memisoglu G, Ang J. Mazella A and others. A pathway of targeted autophagy is induced by DNA damage in budding yeast. Proc Natl Acad Sci U S A. 2017;114(7):E1158–E1167. doi:10.1073/pnas.1614364114.
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G. Riezman H and others. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26(5):663–674. doi:10.1016/j.molcel.2007.04.020.
- Leroy C, Lee SE, Vaze MB, Ochsenbein F, Guerois R, Haber JE, Marsolier-Kergoat MC. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol Cell. 2003;11:827–835.
