ABSTRACT
The reader(s) for acetylated tumor antigen p53 remains elusive. Here, PBRM1 (polybromo-1) is identified as a reader for acetylated lysine382 on p53 through its bromodomain 4 (BD4). PBRM1 BD4 mutants fail to support p53 transcriptional activity and suppress tumor growth. Thus PBRM1 suppresses tumor growth through p53 in kidney cancer.
KEYWORDS:
Cellular tumor antigen p53, encoded by TP53, plays a pivotal role in suppressing tumor growth mainly through its transcriptional activity. Acetylation on p53 lysine residues in p53’s C-terminal domain (CTD) is critical for its transcriptional activity through interaction with other proteins, also known as acetylation ‘readers’. p53 acetylation ‘writers’ such as histone acetyltransferases and ‘erasers’ such as histone deacetylases and sirtuins are well described, but the identities and functions of p53 acetylation ‘readers’ remain unclear.Citation1 CREB-binding protein (CBP) recognizes p53 acetylated at K382 (K382Ac) and enhances p53 transcription.Citation2 SET binds to unacetylated CTD and inhibits p53’s transcriptional activity.Citation3 But they fail to fully account for the biological activity of p53′s acetylated CTD.
Around half of human tumors harbor mutations in TP53. Interestingly, in clear cell Renal Cell Carcinoma (ccRCC), the most common subtype of kidney cancer, only about 3% harbor mutations in TP53.Citation4 So either the p53 pathway is impaired indirectly or it is not important in ccRCC. Polybromo-1 gene (PBRM1) is mutated in approximately 40% of ccRCC tumors.Citation4 PBRM1 (protein polybromo-1) contains six bromodomains (BDs) which are known as readers of acetylated lysines. It is also called BAF180, and it targets a SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeler complex to the chromatins though recognizing acetylated lysine 14 on histone H3 (H3K14Ac) via its BD2.Citation5,Citation6 The components of the SWI/SNF complex have an overall 20% mutation rate in cancers, making it the second most highly mutated entity next to p53. SWI/SNF mutations are also mutually exclusive with TP53 mutations in many cancer types.Citation7 Many SWI/SNF complex components are reported to interact with p53 and are required for p53 function. Notably, PBRM1 is required for p53-mediated replicative senescence in human primary fibroblasts.Citation8 Hence, the authors investigate whether PBRM1, through its BDs, functions as a reader of acetylated p53.
The authors find that PBRM1 binds to p53 in many cell lines. DNA damage, which activates p53, enhances their interaction, whereas deletion of the p53 CTD diminishes it. Increase of p53 CTD acetylation, either by overexpressing p53 acetyltransferases CBP and histone acetyltransferase p300 or treatment with nicotinamide (the inhibitor of sirtuins), enhances the PBRM1-p53 interaction. Moreover, a p53 6KR mutant, in which all six lysine residues in CTD are mutated and cannot be acetylated, fails to bind more PBRM1 when p300 is co-expressed. This suggests that acetylation on the p53 CTD boosts binding to PBRM1.
A series of p53 CTD peptides with or without lysine acetylation(s) are synthesized and assessed for their binding to PBRM1. Only the peptides containing acetylated lysine 382 (K382Ac) of p53 displayed significantly stronger binding to PBRM1 or the PBRM1 BDs than their non-acetylated counterparts. This indicates that BDs of PBRM1 recognize K382Ac on p53.
Among the six BDs on PBRM1, only BD4 shows detectable binding to p53, p53 CTD peptide and the K382Ac peptide. A mutation that disrupts BD4’s ability to recognize acetyl-lysine (BD4*) or two tumor-derived mutations in BD4 identified from cBioPortalCitation9 cancels stronger binding of PBRM1 to the K382Ac peptide over the non-acetylated peptide, suggesting that BD4 of PBRM1 is critical for the recognition of K382Ac on p53.
In H1299 and HCT116 cells which are widely used for p53 function analysis, and in multiple kidney cancer cell lines with wildtype p53, the authors find that loss of PBRM1 reduces the expression of many p53 downstream targets such as p21 (encoded by CDKN1A), Mdm2 and p53 up-regulated modulator of apoptosis (PUMA). Re-expression of wildtype PBRM1, but not the BD4* mutant, rescues the expression of these genes. With chromatin immunoprecipitation (ChIP), the authors find that PBRM1 loss reduces p53 binding to the promoter of CDKN1A.
Further functional analysis reveals that PBRM1 loss compromises G1/S cell cycle arrest after DNA damage. In a nude mouse xenograft model, re-expression of wildtype PBRM1, but not the BD4* mutant, suppresses tumor growth. The tumor suppressor activity is associated with the expression of p53 targets p21 and PUMA. Notably, in kidney cancer patient samples, a significant positive correlation between PBRM1 loss and p21 loss is identified, especially in early tumor grades or stages.
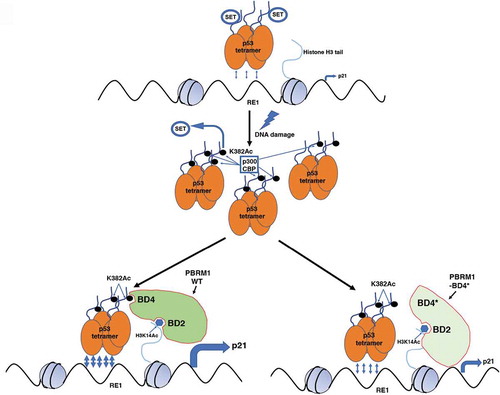
In summary, this study identifies PBRM1 as a functional p53 acetylation reader, elucidates the role of the PBRM1-p53 axis on renal tumor growth, and provides novel insights to the crosstalk between the SWI/SNF complex and the p53 pathway (). It shows that p53 pathway is indirectly impaired in kidney cancer. Since PBRM1 is mutated/disabled in other types of cancers, this finding might have implications beyond renal cancer. In addition, this study provokes new thinking on the interaction between bromodomain and KAc: 1. Different from previous reported p53 peptide of 14 amino acids (aa),Citation5 only the p53 peptide of 26 aa can bind to PBRM1’s BD. This suggests that previous BD screens based on around 15 aa peptides may miss certain interactions; 2. Acetylation on six lysines of p53 CTD including K382 inhibits its binding to PBRM1. This result supports the ‘histone code’ hypothesis and suggests that different combinations of acetylation sites on the p53 CTD can fine-tune p53 transcriptional activity; 3. In agreement with previous report on BD2 and H3K14Ac,Citation6 adjacent BD(s) collaborates with BD4 to create strong binding to p53 K382Ac. Thus the different BDs might work together like fingers to recognize different KAcs.
Figure 1. Potential mechanisms whereby PBRM1 regulates clear cell renal cell carcinoma growth upon recognition of acetylated K382 in p53. SET binds to unacetylated p53 C-terminal domain (CTD) and inhibits p53 transcription. After DNA damage, p53 is acetylated by CREB-binding protein (CBP) or histone acetyltransferase p300. It removes SET binding and provides the K382Ac recognition site for PBRM1 bromodomain 4 (BD4). Wildtype PBRM1 facilitates p53 tetramer binding to response element 1 (RE1) on CDKN1A promoter through the recognition of K382Ac by BD4 and acetylated lysine 14 on histone H3 (H3K14Ac) by BD2. Mutations on BD4 abolished the recognition, reduced p53’s binding to RE1 and compromised p53’s transcription. It is adapted from Cai et. al,Citation10 without any change under the license http://creativecommons.org/licenses/by/4.0/.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011;2:1–3. doi:10.1007/s13238-011-1063-9.
- Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou M-M, et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi:10.1016/S1097-2765(03)00528-8.
- Wang D, Kon N, Lasso G, Jiang L, Leng W, Zhu W-G, Qin J, Honig B, Gu W. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 2016;538:118–122. doi:10.1038/nature19759.
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi:10.1038/nature12222.
- Charlop-Powers Z, Zeng L, Zhang Q, Zhou MM. Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 2010;20:529–538. cr201043[pii]. doi:10.1038/cr.2010.43.
- Liao L, Alicea-Velázquez NL, Langbein L, Niu X, Cai W, Cho E-A, Zhang M, Greer CB, Yan Q, Cosgrove MS, et al. High affinity binding of H3K14ac through collaboration of bromodomains 2, 4 and 5 is critical for the molecular and tumor suppressor functions of PBRM1. Mol Oncol. 2019;13:811–828. doi:10.1002/1878-0261.12434.
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi:10.1038/ng.2628.
- Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci U S A. 2010;107:14280–14285. doi:10.1073/pnas.1009559107.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: figure 1. Cancer Discov. 2012;2:401–404. doi:10.1158/2159-8290.CD-12-0095.
- Cai W, Su L, Liao L, Liu ZZ, Langbein L, Dulaimi E, Testa JR, Uzzo RG, Zhong Z, Jiang W, et al. PBRM1 acts as a p53 lysine-acetylation reader to suppress renal tumor growth. Nat Commun. 2019;10:5800. doi:10.1038/s41467-019-13608-1.
