Abstract
The complete sequence (37 657 bp) of the mitochondrial DNA (mtDNA) of the Saccharina sp. ye-C2 was determined. About 38 protein-coding genes (PCG), 3 ribosomal RNAs (rRNA) and 25 transfer RNA (tRNA) genes were annotated in the genome. The phylogenetic analysis strongly supports the close phylogenetic affinity of Saccharina sp. ye-C2 and Saccharina japonica based on the mitochondrial genomes of other brown algae.
The kelp Saccharina are large seaweeds (algae) belonging to the brown algae (Phaeophyceae), which contain some important economic seaweeds extensively cultivated in China, Japan and Korea (Wang et al. Citation2013). However, close breeding for generations in China caused loss of genetic variance in S. japonica (Zhan et al. Citation2012). Thankfully, the genome of S. japonica has been sequenced (Ye et al. Citation2015), which would powerfully push the genetic improvement of the species. Species and strains of Saccharina all over the world were resequenced in the project mentioned above. Enough Illumina sequencing data were provided to develop more genetic tools for Saccharina. In this study, the complete mitochondrial genome of a wild strain (NO. KT336420), sampled in Liaoning Province and named Saccharina sp. ye-C2 based on the phylogenetic analysis with 16 complete brown algae mitochondrial genomes.
The length of complete of Saccharina sp. ye-C2 is 37 657 bp and the genome contains 38 protein-coding genes (rps2-4, rps7-8, rps10-14, rps19, atp6, atp8, atp9, cox1-3, nad1-7, nad9, nad11, nad4L, rpl2, rpl5, rpl6, rpl14, rpl16, rpl31, ORF41, ORF130, ORF377, tatC and cob), 25 transfer RNA (tRNA) genes and 3 ribosomal RNA (rRNA) genes (5S rRNA, 16S rRNA and 23S rRNA). All 38 protein-coding genes (PCGs) have typical initiation codons (ATG). The numbers of PCGs that have complete termination codons TAA, TAG, TGA are 26, 8 and 4, respectively. Nucleotide frequency of the H-strand is as follows: T, 36.29%; A, 28.41%; C, 14.72% and G, 20.58%. The mitogenome of Saccharina sp. ye-C2 encodes 9637 amino acids, excluding the stop codons. All the 25 typical tRNAs, ranging from 71 to 88, possess a complete clover leaf secondary structure. The rRNAs of the 5S rRNA, 16S rRNA and 23S rRNA genes are 133, 1535 and 2745 bp in length, respectively.
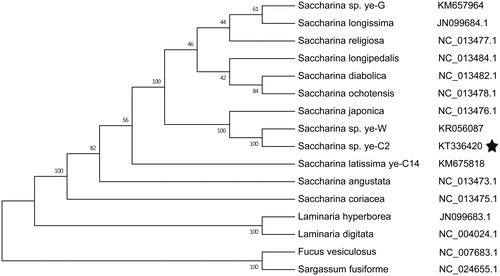
Phylogenetic analysis based on other 16 brown algae complete mitochondrial sequence data show that Saccharina sp. ye-C2 belongs to a Saccharina clade and are closely related to S. japonica (). This result was consistent with recent phylogenetic analyses and certain morphological characters (Zhang et al. Citation2013; Guan et al. Citation2014).
Declaration of interest
This work was supported by Scientific Research Funds for Central Non-profit Institutes, Yellow Sea Fisheries Research Institute (20603022015004), National Basic Research Special Foundation of China (2013FY110700) and the Science Fund for Distinguished Young Scholars of Shandong Province (JQ201509) the program of leading talents of Qingdao (13-CX-27). The authors report no conflicts of interest.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402.
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Greco M, Sáez CA, Brown MT, Bitonti MB. 2014. A simple and effective method for high quality co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the Model Brown Alga. PLoS One 9:e96470.
- Guan Z, Fan X, Wang S, Xu D, Zhang X, Wang D, Miao Y, Ye N. 2014. Sequencing of complete mitochondrial genome of brown algal Saccharina sp. ye-G[J]. Mitochondrial DNA. 2014:1–2. doi: 10.3109/19401736.2014.982588.
- Wang WJ, Wang FJ, Sun XT, Liu FL, Liang ZR. 2013. Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae). Planta 237:1123–1133.
- Ye N, Zhang X, Miao M, Fan X, Zheng Y, Dong X, Wang J, Zou L, Wang D, Gao Y, et al. 2015. Saccharina genomes provide novel insight into kelp biology[J]. Nat Commun. 6. doi:10.1038/ncomms7986.
- Zhan X, Fan FL, You WW, Yu JJ, Ke CH. 2012. Construction of an integrated map of Haliotis diversicolor using microsatellite markers. Mar Biotechnol. 14:79–86.
- Zhang J, Wang X, Liu C, Jin Y, Liu T. 2013. The complete mitochondrial genomes of two brown algae (Laminariales, Phaeophyceae) and phylogenetic analysis within Laminaria. J Appl Phycol. 251247–1253.