Abstract
The total length of mitochondrial genome of Lanius schach is 16 820 bp and contains 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and 1 D-loop. The phylogenetic tree of L. schach and 12 other closely species belonging to order Passeriformes was built.
The Lanius is the largest genus in the family Laniidae, contains 29 species (Gill & Donsker Citation2015). Only three species’ mitochondrial genomes of this genus had been reported (Qian et al. Citation2013; Liu et al. Citation2015a, Citation2015b). The Lanius schach, a partial migrant, is widely distributed in Asia (MacKinnon & Phillipps Citation2000). In this article, we determined and described the complete mitogenome of L. schach in order to obtain basic genetic information about this species.
The specimen of L. schach was collected from Xiuning County, Anhui Province, China (29.45°N, 118.15°E) and deposited in the Museum of Huangshan University (Voucher number: HS15003). The complete mitogenome of L. schach (Genbank accession number KU058639) was sequenced to be 16 820 bp which consisted of 13 typical vertebrate protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and 1 D-loop. The overall base composition of the entire genome was as follows: A (31.1%), T (25.7%), C (28.5%) and G (14.7%). All the 13 protein-coding genes initiate with ATG as start codon, except for COI, which begin with GTG. Nine protein-coding genes end with complete stop codons (TAA, AGG and AGA), and the other four genes terminate with T as the incomplete stop codons, which were presumably completed as TAA by post-transcriptional polyadenylation (Anderson et al. Citation1981). Among the mitochondrial protein-coding genes, the ATP8 was the shortest, while the ND5 was the longest. The 22 tRNA genes range in size from 66 to 75 bp. The lengths of 12S and 16S rRNA are 976 bp and 1598 bp. The D-loop in size is 1245 bp.
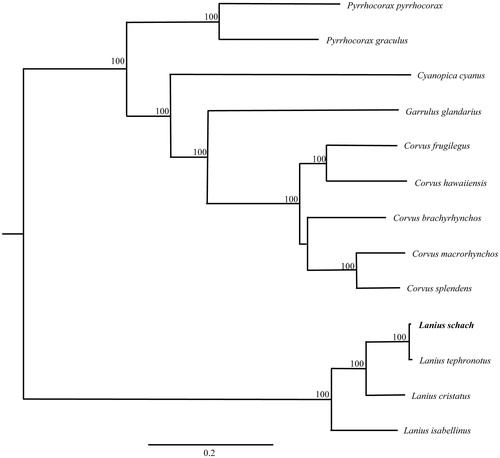
We selected the 12 protein-coding genes located on heavy strand except for ND6 which encoded on the light strand of L. schach, and together with other 12 related species to perform phylogenetic analysis. A maximum-likelihood tree was constructed by online tool RAxML (Stamatakis et al. Citation2008) (). The phylogenetic analysis result was consistent with the previous researches.
Figure 1. A maximum-likelihood (ML) tree of the 13 species from Passeriformes was constructed based on the dataset of 12 concatenated mitochondrial protein-coding genes by online tool RAxML. The numbers above the branch meant bootstrap value. Bold black branches highlighted the study species and corresponding phylogenetic classification. The analyzed species and corresponding NCBI accession number as follows: Pyrrhocorax graculus (KJ598623), Pyrrhocorax pyrrhocorax (KJ598622), Cyanopica cyanus (JN108020), Garrulus glandarius (JN018413), Corvus frugilegus (Y18522), Corvus hawaiiensis (KP161620), Corvus brachyrhynchos (KP403809), Corvus macrorhynchos (KR072661), Corvus splendens (KJ766304), Lanius schach (KU058639), Lanius tephronotus (JX486029), Lanius cristatus (KT004451), Lanius isabellinus (KP995437).
Acknowledgements
We thank Dr. Song Huang (Huangshan University) for valuable comments on this manuscript.
Declaration of interest
This project was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290:457–465.
- Gill F, Donsker D. 2015. IOC World Bird List (v 5.4); [cited 2015 Oct 24]. Available from: http://www.worldbirdnames.org.
- Liu FQ, Bao XK, Fan YN, Li JD. 2015a. Sequencing and analysis of the complete mitochondrial genome of Rufous-tailed Shrike, Lanius isabellinus (Passeriformes, Laniidae). Mitochondrial DNA [Epub ahead of print].
- Liu FQ, Bao XK, Fan YN, Li JD, Yao XX. 2015b. Sequencing and analysis of the complete mitochondrial genome of Brown Shrike, Lanius cristatus (Passeriformes, Laniidae). Mitochondrial DNA [Epub ahead of print].
- MacKinnon J, Phillipps K. 2000. A field guide to the birds of China. London: Oxford University Press.
- Qian CJ, Yan X, Guo ZC, Wang YX, Li XX, Yang JK, Kan XZ. 2013. Characterization of the complete mitochondrial genome of the grey-backed Shrike, Lanius tephronotus (Aves: Passeriformes): the first representative of the family Laniidae with a novel CAA stop codon at the end of cox2 gene. Mitochondrial DNA 24:359–361.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 75:758–771.
