Abstract
The Gekkonidae is the second-most-numerous Family in Sauria and widely distributed around the world. In this paper, the complete mitochondrial genomes of two gecko species, Gekko hokouensis and Gekko japonicas, were sequenced. The lengths of the two mitochondrial genomes (G. hokouensis and G. japonicus) are 17 956 bp and 17 769 bp, respectively. These mitochondrial genomes both contain 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes and a non-coding region (control region). The overall base compositions of the H-strand of G. hokouensis and G. japonicus are 32.1% A, 26.7% T, 27.0% C, 14.2% G and 31.6% A, 25.2% T, 28.6% C, 14.7% G, respectively. Phylogenetic analysis showed that G. hokouensis is a sister clade with G. swinhonis. G. japonicus has a close phylogenetic relationship to the clade of G. hokouensis and G. swinhonis.
Sixteen species of genus Gekko (Squamata: Gekkonidae) is distributed in China (Cai et al. Citation2015). Four complete mitochondrial genomes of Gekko are available in GenBank data (Zhou et al. Citation2006; Qin et al. Citation2011; Li et al. Citation2013; Hao et al. Citation2015). To analyze the mt genomes of other Gekko species, we sequenced the mt genomes of G. hokouensis and G. japonicus collected from Luxi Island (27°59′N, 121°10′) and Chashan (27°55′N, 120°42′), Wenzhou, Zhejiang, China, respectively. Voucher specimens were deposited in Wenzhou University. The accession numbers of G. hokouensis and G. japonicus are KT005801 and KT005800 in GenBank, respectively.
The lengths of complete mitochondrial genomes of G. hokouensis and G. japonicus were 17 769 bp and 17 956 bp, respectively. Both mitogenomes encode 37 genes including 13 respiratory protein-coding genes, two rRNA genes and 22 tRNA genes and the major non-coding region. The gene arrangement structure and transcribing directions were identical to other geckos. The canonical cloverleaf secondary structures predicted by tRNAscan-SE online server (http://lowelab.ucsc.edu/tRNAscan-SE/) could be found in all tRNA genes except tRNAPhe, tRNACys and one tRNASer. tRNASer lacked dihydrouridine (DHU) arm, which was a common feature observed in vertebrate animals (Meganathan et al. Citation2011).
Twelve protein-coding genes, 14 tRNAs and two rRNAs are transcribed from the heavy strand (H-strand), whereas ND6 and the other eight tRNAs are located on the light strand (L-strand). The overall base compositions of the H-strand of G. hokouensis and G. japonicus are 32.1% A, 26.7% T, 27.0% C, 14.2% G and 31.6% A, 25.2% T, 28.6% C, 14.7% G, respectively. The overall A + T content of mt genomes are 58.8% for G. hokouensis and 56.8% for G. japonicus. The overall AT and GC skews for G. hokouensis and G. japonicus are 0.091, −0.312 and 0.112, −0.321, respectively. The 12S and 16S rRNA genes of the G. hokouensis and G. japonicus are located between tRNAPhe and tRNALeu genes, and separated by tRNAVal gene. The control regions of G. hokouensis and G. japonicus located between the tRNAPhe and tRNAPro genes are 2390 bp and 2539 bp in length with the A + T content of 63.0% and 64.6%, respectively.
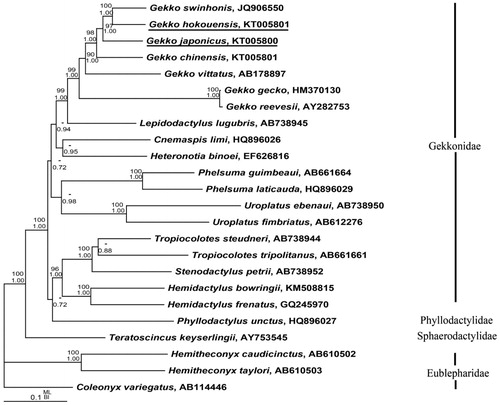
To elucidate the phylogenetic relationships of gekkonids, 22 sequences of the complete (or nearly complete) mitochondrial genomes were obtained from GenBank database. Bayesian inference (BI) and maximum likelihood (ML) trees were analyzed by using MrBayes 3.1.2 (Huelsenbeck & Ronquist Citation2001) and PAUP* 4.0b10 (Swofford Citation2002) (). Coleonyx variegatus, Hemitheconyx caudicinctus and H. taylori were selected as outgroups. Phylogenetic analysis showed that G. hokouensis was a sister clade with G. swinhonis and the clade of G. hokouensis and G. swinhonis was a sister with G. japonicus. The monophyly of Gekko is well supported, but the monophyly of Gekkonidae is failed for Phyllodactylus unctus (Phyllodactylidae) within Gekkonidae, which is inconformity to the results of Pyron et al. (Citation2013). More species of Phyllodactylidae are need to further study the relationship of Gekkonidae and Phyllodactylidae.
Figure 1. Bayesian inference (BI) and maximum likelihood (ML) phylogeny results of the 24 species from 13 mt protein-coding genes were analyzed. The nodal support above branches is shown as posterior probabilities from BI and bootstrap percentages from ML. Branch lengths and topology are from the Bayesian analysis.
Acknowledgements
We are deeply indebted to Dr. Jiayong Zhang from College of Chemistry and Life Science, Zhejiang Normal University, China, for his kindness in helping our work by giving some advice about the paper. Particular thanks must go to Prof. Rongquan Zheng of the Institute of Ecology, Zhejiang Normal University, for giving the instruments for the experiment.
Declaration of interest
This work was supported by the grants from the National Natural Science Foundation of China (31170376). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Cai B, Wang YZ, Chen YY, Li JT. 2015. A revised taxonomy for Chinese reptiles. Biodivers Sci. 23:365–382.
- Hao SL, Ping J, Zhang YP. 2015. Complete mitochondrial genome of Gekko chinensis (Squamata, Gekkonidae). Mitochondrial DNA. Early Online: 1–2. DOI: 10.3109/19401736.2015.1022751.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755.
- Li HM, Zeng DL, Guan QX, Qin PS, Qin XM. 2013. Complete mitochondrial genome of Gekko swinhonis (Squamata, Gekkonidae). Mitochondrial DNA 24:86–88.
- Meganathan PR, Dubey B, Batzer MA, Ray DA, Haque I. 2011. Complete mitochondrial genome sequences of three Crocodylus species and their comparison within the Order Crocodylia. Gene 478:35–41.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:93.
- Qin XM, Qian F, Zeng DL, Liu XC, Li HM. 2011. Complete mitochondrial genome of the red-spotted tokay gecko (Gekko gecko, Reptilia: Gekkonidae): comparison of red and black-spotted tokay geckos. Mitochondrial DNA 22:176–177.
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sunderland, MA: Sinauer Associates.
- Zhou KY, Li HD, Han DM, Bauer AM, Feng JY. 2006. The complete mitochondrial genome of Gekko gecko (Reptilia: Gekkonidae) and support for the monophyly of Sauria including Amphisbaenia. Mol Phylogenet Evol. 40:887–892.
