Abstract
We present the complete mitochondrial genome of Cipangopaludina cathayensis in this study. The mitochondrial genome is 15 706 bp in length, containing 13 protein-coding genes, two rRNA genes and 22 tRNA genes. Overall nucleotide compositions of the light strand are 40.97% of A, 30.78% of T, 20.48% of C and 12.60% of G. Its gene arrangement and distribution are different from the typical vertebrates. The absence of D-loop is consistent with the Gastropoda, but, at least, one lengthy non-coding region is an essential regulatory element for the initiation of transcription and replication. A phylogenetic tree is constructed using the maximum-likelihood method based on the complete mitogenomes of the closely related 21 Gastropoda species to assess their actual phylogenetic relationship and evolution. The result provides fundamental data for resolving phylogenetic and genetic problems related to effective management strategies.
The golden apple snail Pomacea canaliculata (Lamarck 1822) indigenous to South America, was introduced from Argentina to Taiwan for commercial purposes, subsequently to numerous countries throughout southern and eastern Asia including China, becoming pests of wetland rice and other crops and causing massive economic losses. They also continue to spread into non-agricultural wetlands and their ecological impacts are more difficult to estimate (Wood et al. Citation2005; Levin et al. Citation2006; Rawlings et al. Citation2007). The United States Aquatic Nuisance Species Task Force listed P. canaliculata among the world’s 100 worst invasive species (Lowe et al. Citation2000). But, the confused taxonomy and difficult identification of genus Pomacea results from the overall highly conserved external morphology across the genus yet considerable intraspecific shell variation, obscuring the true number of species and their identities (Thiengo et al. Citation1993; Cazzaniga Citation2002; Cowie Citation2002; Cowie et al. Citation2006).
We sequenced its complete mitogenome to analyze phylogenetic relationship and evolutionary history for broader understanding of invasion processes and implementing effective management strategies. The specimen was sampled from Ningxi Teaching Experimental Base of South China Agricultural University in Guangzhou (E 113°29′, N 23°5′), and stored in the specimen museum of SCAU (accession number: 201502116).
The complete mitochondrial genome of P. canaliculata (GenBank accession number KU052865) is 15 706 bp in length, containing 13 protein-coding genes, two ribosomal RNA genes (L-rRNA and S-rRNA), 22 transfer RNA genes (tRNA). The rest of them are encoded on the heavy strand except eight tRNA genes (Met, Tyr, Cys, Trp, Gln, Gly, Glu, Thr) on the light strand. Twenty-two tRNA genes vary from 62 to 70 bp in length, and all fold into the typical cloverleaf secondary structure. Among 13 protein-coding genes (total 11 220 bp) encoding 3727 amino acids, the maximum is ND5 with 1710 bp and the minimum is ATP8 with only 159 bp. S-rRNA and L-rRNA genes are 877 and 1381 bp, respectively, located between the tRNAGlu and tRNALeu genes and separated by the tRNAVal gene. Overall nucleotide compositions of the light strand in descending order are 40.97% of A, 30.78% of T, 20.48% of C and 12.60% of G. Gene arrangement and distribution are different from the typical vertebrates (Yang et al. Citation2014a, Citation2016a–g) and similar to Cipangopaludina cathayensis (Yang et al. Citation2014b). The absence of D-loop is consistent with the Gastropoda (Liu et al. Citation2012; Zeng et al. Citation2015; Zhou et al. Citation2016), but, at least, one lengthy non-coding region is an essential regulatory element for the initiation of transcription and replication (Wolstenholme Citation1992).
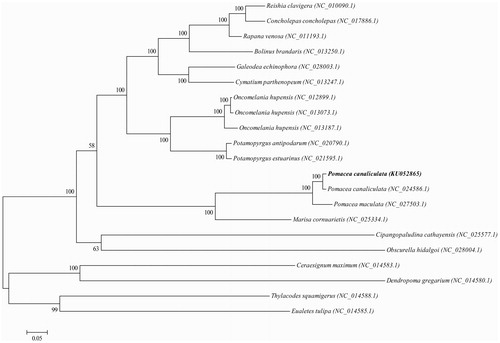
A phylogenetic tree is constructed using the maximum-likelihood method based on the complete mitogenomes of the closely related 21 Gastropoda species to assess their actual phylogenetic relationship and evolution (). But repeat elements (AAAGAAACTAAGAGATAAGATAT)N and (AGTTTCTTTATATCTTATCTCTT)N which are located between tRNA-Phe (GAA) and COX3 gene pair into the complex hairpin structure to prevent the PCR process. So, it is difficult to verify the accurate information by Hiseq2500 sequencing system and needs further research.
Figure 1. Phylogenetic tree generated by the maximum-likelihood method based on the complete mitochondrial genomes. The published sequences in GenBank adopted are Reishia clavigera (NC_010090.1), Concholepas concholepas (NC_017886.1), Rapana venosa (NC_011193.1), Bolinus brandaris (NC_013250.1), Galeodea echinophora (NC_028003.1), Cymatium parthenopeum (NC_013247.1), Oncomelania hupensis (NC_012899.1), Oncomelania hupensis (NC_013073.1), Oncomelania hupensis (NC_013187.1), Potamopyrgus antipodarum (NC_020790.1), Potamopyrgus estuarinus (NC_021595.1), Pomacea canaliculata (KU052865), Pomacea canaliculata (NC_024586.1), Pomacea maculata (NC_027503.1), Marisa cornuarietis (NC_025334.1), Cipangopaludina cathayensis (NC_025577.1), Obscurella hidalgoi (NC_028004.1), Ceraesignum maximum (NC_014583.1), Dendropoma gregarium (NC_014580.1), Thylacodes squamigerus (NC_014588.1), Eualetes tulipa (NC_014585.1).
Declaration of interest
This work is supported by the National Natural Science Foundation of China (No. U1131006, No. 30770403, No. 30900187 and No. 31502144), Guangdong Science and Technology Program (No. 2007B02079007, No. 2011B020309009 and No. 2013B020308001), Research and Development Projects of Marine Fishery Science and Technology in Guangdong Province (No. A201501A09), the Natural Science Foundation of Guangdong Province (No. 2015A030313409), Foundation for High-level Talents in Higher Education of Guangdong Province. The authors report no conflicts of interest. The authors themselves are responsible for the content and writing of the paper.
References
- Cazzaniga NJ. 2002. Old species and new concepts in the taxonomy of Pomacea (Gastropoda: Ampullariidae). Biocell 26:71–81.
- Cowie RH. 2002. Apple snails (Ampullariidae) as agricultural pests: their biology, impacts and management. In: Barker GM, editor. Molluscs as crop pests. Wallingford, UK: CAB-International. p. 145–192.
- Cowie RH, Hayes KA, Thiengo SC. 2006. What are apple snails? Confused taxonomy and some preliminary resolution. In: Joshi RC, Sebastian LS, editors. Global advances in ecology and management of golden apple snails. Nueva Ecija, Philippines: Philippine Rice Research Institute. p. 3–24.
- Levin P, Cowie RH, Taylor JM, Hayes KA, Burnett KM, Ferguson CA. 2006. Apple snail invasions and the slow road to control: ecological, economic, agricultural and cultural perspectives in Hawaii. In: Joshi RC, Sebastian LS, editors. Global advances in ecology and management of golden apple snails. Nueva Ecija, Philippines: Philippine Rice Research Institute. p. 325–335.
- Liu GH, Wang SY, Huang WY, Zhao GH, Wei SJ, Song HQ, Xu MJ, Lin RQ, Zhou DH, Zhu XQ. 2012. The complete mitochondrial genome of Galba pervia (Gastropoda: Mollusca), an intermediate host snail of Fasciola spp. PLoS One 7:e42172.
- Lowe S, Browne M, Boudjelas S, De Poorter M. 2000. 100 of the world’s worst invasive alien species: a selection from the global invasive species database. Auckland, New Zealand: The Invasive Species Specialists Group of the Species Survival Commission of the World Conservation Union.
- Rawlings TA, Hayes KA, Cowie RH, Collins TM. 2007. The identity, distribution, and impacts of non-native apple snails in the Continental United States. BMC Evol Biol. 7:97.
- Thiengo SC, Borda CE, Barros Araújo JL. 1993. On Pomacea canaliculata (Lamarck, 1822) (Mollusca; Pilidae: Ampullariidae). Mem I Oswaldo Cruz. 88:67–71.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Wood T, Anurakpongsatorn P, Chaichana R, Mahujchariyawong J, Satapanajaru T. 2005. Predation on freshwater bryozoans by the apple snail, Pomacea canaliculata, Ampulariidae [sic], an invasive species in Southeast Asia: a summary report. Denisia 16:283–286.
- Yang H, Sun J, Zhao H, Chen Y, Yang Z, Li G, Liu L. 2016e. The complete mitochondrial genome of the Clarias fuscus (Siluriformes, Clariidae). Mitochondrial DNA 27(2):1255–6.
- Yang H, Sun J, Zhao H, Yang Z, Xiao S, Li G, Liu L. 2016f. The complete mitochondrial genome of Nibea coibor (Perciformes, Sciaenidae). Mitochondrial DNA 27(2):1520–2.
- Yang H, Xie Z, Li S, Wu X, Peng C, Zhang Y, Lin H. 2014a. The complete mitochondrial genome of the orange-spotted grouper Epinephelus coioides (Perciformes, Serranidae). Mitochondrial DNA.
- Yang H, Zhang JE, Luo H, Luo M, Guo J, Deng Z, Zhao B. 2014b. The complete mitochondrial genome of the mudsnail Cipangopaludina cathayensis (Gastropoda: Viviparidae). Mitochondrial DNA.
- Yang H, Zhao H, Sun J, Chen Y, Liu L, Zhang Y, Liu L. 2016d. The complete mitochondrial genome of the Hemibarbus medius (Cypriniformes, Cyprinidae). Mitochondrial DNA 27(2):1070–2.
- Yang H, Zhao H, Sun J, Xie Z, Yang Z, Liu L. 2016b. The complete mitochondrial genome of the Culter recurviceps (Teleostei, Cyprinidae). Mitochondrial DNA 27(1):762–3.
- Yang H, Zhao H, Sun J, Yang Z, Xiao S, Li G, Liu L. 2016g. The complete mitochondrial genome of the amoy croaker Argyrosomus amoyensis (Perciformes, Sciaenidae). Mitochondrial DNA 27(2):1530–2.
- Yang H, Zhao H, Sun J, Zhang Y, Yang Z, Liu L. 2016c. The complete mitochondrial genome of the Hemibagrus wyckioides (Siluriformes, Bagridae). Mitochondrial DNA 27(1):766–8.
- Yang H, Zhao H, Xie Z, Sun J, Yang Z, Liu L. 2016a. The complete mitochondrial genome of the Hemibagrus guttatus (Teleostei, Bagridae). Mitochondrial DNA 27(1):679–81.
- Zeng T, Yin W, Xia R, Fu C, Jin B. 2015. Complete mitochondrial genome of a freshwater snail, Semisulcospira libertina (Cerithioidea: Semisulcospiridae). Mitochondrial DNA 26(6):897–8.
- Zhou X, Chen Y, Zhu S, Xu H, Liu Y, Chen L. 2016. The complete mitochondrial genome of Pomacea canaliculata (Gastropoda: Ampullariidae). Mitochondrial DNA 27(2):884–5.
