Abstract
Macropodus erythropterus is a small well-known aquarium fish. In the present study, the complete mitochondrial DNA (mtDNA) sequences of M. erythropterus were first determined. The mtDNA of M. erythropterus (GenBank accession no. KU215670) was a circular molecule of 16 495 bp in length with two ribosome RNA (rRNA) genes, 13 protein-coding genes, 22 transfer RNA genes, an L-strand replication origin and a control region (CR). The entire mitogenome nucleotide acid was 15.71% for G, 29.66% for A, 28.37% for T and 26.26% for C with an A + T content of 58.03%. And the A + T contents of 12S rRNA, 16S rRNA and CR were 55.63%, 57.66% and 66.79%, respectively. This study provides basic molecular data for studying the conservation biology, phylogenetic and evolutionary analysis of Macropodusinae fishes.
Macropodus erythropterus belongs to the family Osphronemidae and the order Perciformes. In nature, M. erythropterus is distributed in China and Viet Nam, but it has been widely transported into many countries as an ornamental fish, especially in Germany. Macropodus fishes are so fascinating that they were given a name of paradise fish in the Western countries. However, the taxonomy of M. erythropterus was controversial. The name of M. erythropterus was first given according to its body colour by Freyhof and Herder in Citation2002 for the distinction with Macropodus spechti. But Winstanley and Clements’s (Citation2008) research showed that it cannot differentiate M. erythropterus and M. spechti through morphology methods. In this study, the complete mitochondrial genome of M. erythropterus was determined to provide basic molecular data for studying its conservation biology, phylogenetic and evolutionary analysis.
The test fish was collected from ornamental fish market in Huizhou, Guangdong Province, China and total DNA from muscles was extracted using the Ezup Column Animal Genomic DNA Kit (Sangon, Shanghai, China). The specimen was numbered as 201509ME1 and stored in the specimen storeroom of College of Animal Science and Technology, Anhui Agricultural University, Hefei, P.R. China. The primers were initially designed based on members of the Macropodus fishes: Macropodus opercularis (GenBank accession no. KM588227.1) and Macropodus ocellatus (GenBank accession no. KJ813282.1). DNA sequences were analyzed using the DNAstar software (DNAStar, Madison, WI, USA).
The M. erythropterus mitochondrial genome (GenBank accession no. KU215670) was a circular molecule of 16 495 bp in length with 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosome RNA (rRNA) genes, an L-strand replication origin and a control region (CR). The entire mitogenome nucleotide acid was 15.71% for G, 29.66% for A, 28.37% for T and 26.26% for C with an A + T content of 58.03%.
Like other vertebrates (Pereira Citation2000), mitochondrial genes of M. erythropterus were encoded on the heavy strand (H) except for ND6 and eight tRNA genes. Two start codons (ATG and GTG) and three stop codons (TAA, TA and T) were found during the 13 PCGs. 12 of PCGs began with ATG start codon and the rest one (COX1) started with GTG start codon, typically in all bony fishes (Li Citation2012). Six PCGs (ND1, ATP8, ATP6, ND4L, ND5 and ND6) harboured the complete termination codon: TAA, and seven PCGs had an incomplete termination codon: TA (ND2, COX1, COX3 and CYTB) or T (COX2, ND3 and ND4), which were presumably completed as a stop codon TAA by polyadenylation (Ojala et al. Citation1981).
Twenty-two tRNA genes were interspersed between PCGs and rRNA genes, ranging in size from 66 bp to 74 bp. The A + T contents of 12S rRNA, 16S rRNA and CR were 55.63%, 57.66% and 66.79%, respectively.
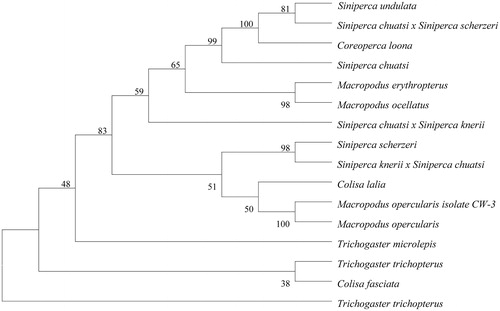
The MP tree made by MEGA version 5.1 programme (Tamura et al. Citation2011) was constructed on the basis of the complete mitochondrial DNA from M. erythropterus and other 15 of the most closely related fish species in the GenBank database ().
Figure 1. MP tree was generated using the MEGA version 5.1 programme. Numbers on the nodes correspond to bootstrap values based on 1000 iterations. The accession numbers of the species are Macropodus opercularis isolate CW-3 (KP234027.1), Macropodus opercularis (KM588227.1), Macropodus ocellatus (KJ813282.1), Trichogaster microlepis (KP342379.1), Trichogaster trichopterus (KP100265.1), Trichogaster trichopterus (LC026155.1), Colisa lalia (AP006039.1), Colisa fasciata (KP301136.1), Siniperca scherzeri (AP014527.1), Siniperca undulate (KJ644783.1), Siniperca chuatsi × Siniperca scherzeri (KJ907732.1), Siniperca chuatsi (JF972568.1), Siniperca chuatsi × Siniperca knerii (KJ960194.1), Siniperca knerii × Siniperca chuatsi (KM024309.1), Coreoperca loona (KJ644781.1).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Freyhof J, Herder F. 2002. Review of the paradise fishes of the genus Macropodus in Vietnam, with description of two new species from Vietnam and southern China (Perciformes: Osphronemidae). Ichthyol Explor Freshwaters 13:147–168.
- Li S-J. 2012. The complete mitochondrial genomes of largemouth bass of the northern subspecies (Micropterus salmoides salmoides) and Florida subspecies (Micropterus salmoides floridanus) and their applications in the identification of largemouth bass species. Mitchondrial DNA 23:92–99.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474.
- Pereira SL. 2000. Mitochondrial genome organization and verte-brate phylogenetics. Genet Mol Biol. 23:745–752.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011 MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739
- Winstanley T, Clements KD. 2008. Morphological re-examination and taxonomy of the genus Macropodus (Perciformes, Osphronemidae). Zootaxa 19:1–27.
