Abstract
The complete mitochondrial genome of the Ukrainian nine-spined stickleback Pungitius platygaster was obtained using massive parallel sequencing of genomic DNA. The mitogenome sequence was 16 570 bp long, and the gene order and contents were identical to those of other sequenced Pungitius mitogenomes. In a phylogenetic analysis, the mitogenome of P. platygaster clustered with other Pungitius mitogenomes, yet being clearly distinct from those of P. pungitius, P. sinensis and P. kaibarae.
The Ukrainian nine-spined stickleback P. platygaster is morphologically one of the most easily identifiable sticklebacks in the genus Pungitius (Keivany & Nelson Citation2000). Hence, most taxonomic treatments recognize it as a valid species, rather than a subspecies of P. pungitius (Wootton Citation1984; Keivany & Nelson Citation2000; Mattern Citation2007). However, in contrast to most other species in this genus, the genetics of this species is poorly studied. To the best of our knowledge, the only genetic studies of this species involved phylogenetic analysis of mitochondrial DNA (mtDNA) fragments by Geiger et al. (Citation2014) and Wang et al. (Citation2015). Access to the complete mitogenome of this species might be helpful in piecing together the evolutionary affinities and biogeographic history of this species which (together with P. hellenicus) has an enigmatic southern distribution at the Black Sea, Sea of Azov, Aral Sea and Caspian Sea basins. All other members of the genus have much more northern distribution ranges (Wootton Citation1976, Citation1984).
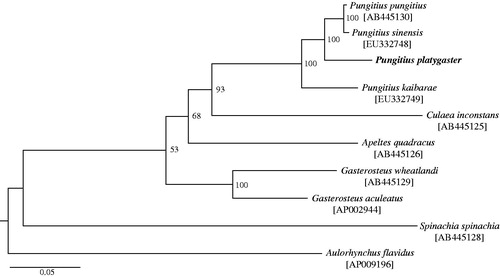
We sequenced the genomic DNA of P. platygaster collected from the River Loudias drainage in Greece (40°50′N, 22°18'E) on the Illumina HiSeq2000 platform with 100 paired-end strategy. A total of 11.8 million reads were aligned against the P. sinensis mitogenome (Hwang et al. Citation2012a) with bwa-0.5.10 (Li & Durbin Citation2009). Mean sequence coverage across the genome was 27.58 reads per base pair and 100% of the reference genome had one-fold coverage (72.61% of the bases had ≥20-fold coverage). The consensus sequence of the P. platygaster mitochondrial genome was exported with samtools 1.2 (Li et al. Citation2009) and manually checked. The complete mitochondrial genome of P. platygaster is 16 570 bp (GenBank Accession No. KT989570), consisting of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and a control region. The order and direction of these genes were identical to those of other Gasterosteidae fishes (Miya et al. Citation2001; Kawahara et al. Citation2009; Hwang et al. Citation2012a, Citation2012b). Of the 13 protein-coding genes, four (ND2, COII, ND4 and Cytb) showed an incomplete stop codon. The base composition of the entire mitochondrial genome was 27.8% for A, 26.6% for T, 17.2% for G and 28.4% for C. The phylogenetic position of P. platygaster in Gasterosteidae fishes was investigated based on a maximum likelihood tree constructed with the 37 genes (15 583 bp in total) using RAxML v.8.0 under the GTR + GAMMA model (37 gene partitions and 100 thorough bootstrap replicates; Stamatakis Citation2014). P. platygaster was phylogenetically positioned with other Pungitius species (i.e., P. pungitius, P. sinensis and P. kaibarae), but showed clear albeit shallow divergence from them (). Nucleotide identity in the 37 gene regions of P. platygaster was 95.7%, 95.6% and 93.0% in comparison with those of P. pungitius (GenBank Accession No. AB445130; Kawahara et al. Citation2009), P. sinensis (EU332748; Hwang et al. Citation2012a) and P. kaibarae (EU332749; Hwang et al. Citation2012b), respectively.
Acknowledgements
We thank Jacquelin De Faveri for linguistic corrections. Pekka Ellonen, Laura Häkkinen, Tiina Hannunen and Sami Karja are thanked for help with laboratory and bioinformatic work. The sequencing was conducted at the Finnish Institute for Molecular Medicine.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by grants (108601 & 118673) from the Academy of Finland, the Ministry of Culture of the Czech Republic (DKRVO 2015/15, National Museum, 00023272) and by the institutional resources of the Ministry of Education, Youth, and Sports of the Czech Republic.
References
- Geiger MF, Herder F, Monaghan MT, Almada V, Barbieri R, Bariche M, Berrebi P, Bohlen J, Casal-Lopez M, Delmastro GB, et al. 2014. Spatial heterogeneity in the Mediterranean Biodiversity Hotspot affects barcoding accuracy of its freshwater fishes. Mol Ecol Resour. 14:1210–1221.
- Hwang D-S, Song HB, Lee J-S. 2012a. Complete mitochondrial genome of the Chinese stickleback Pungitius sinensis (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:293–294.
- Hwang D-S, Song HB, Lee J-S. 2012b. Complete mitochondrial genome of the Amur stickleback Pungitius kaibarae (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:313–314.
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M. 2009. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 50:401–404.
- Keivany Y, Nelson JS. 2000. Taxonomic review of the genus Pungitius, ninespine sticklebacks (Gasterosteidae). Cybium 24:107–122.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079.
- Mattern MY. 2007. Phylogeny, systematics and taxonomy of sticklebacks. In: Östlund-Nilsson S, Mayer I, Huntingford FA., editors. Biology of the three-spined stickleback. Boca Raton: CRC Press. p. 1–40.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenetic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Phylogenet Evol. 18:1993–2009.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313.
- Wang C, Shikano T, Persat H, Merilä J. 2015. Mitochondrial phylogeography and cryptic divergence in the stickleback genus Pungitius. J Biogeogr 42:2334–2348.
- Wootton RJ. (1976). The Biology of the Sticklebacks. Academic Press, New York.
- Wootton RJ. (1984). A Functional Biology of Sticklebacks. Croom Helm, London.