Abstract
The complete mitochondrial genome sequences of three divergent lineages of the smooth tail nine-spined stickleback (Pungitius laevis) were obtained with massive parallel sequencing of their genomic DNA. The genome sequences were 16 574–16 580 bp long, and the gene order and contents were identical to those of other sequenced Pungitius mitogenomes. Although the mitogenome sequences of all three P. laevis lineages clustered within the genus Pungitius, they were clearly distinct and showed divergence comparable to that seen between some Pungitius species.
The taxonomic status and phylogenetic positioning of the smooth tail nine-spined stickleback Pungitius laevis has been the subject of considerable debate over the past decades (Münzing Citation1969; Gross Citation1979; Keivany & Nelson Citation2000). While some treatments considered it to be a subspecies of the nine-spined stickleback P. pungitius (Keivany & Nelson Citation2000), others have classified it as a taxonomically valid and independent species (Kottelat & Freyhof Citation2007). A recent study based on partial mitochondrial DNA (mtDNA) fragments revealed the existence of three highly distinct mtDNA lineages within P. laevis and suggested that they have diverged 1.4–2.0 Mya (Wang et al. Citation2015). Access to full mtDNA should help to further clarify the evolutionary history and phylogenetic affinities of these P. laevis lineages.
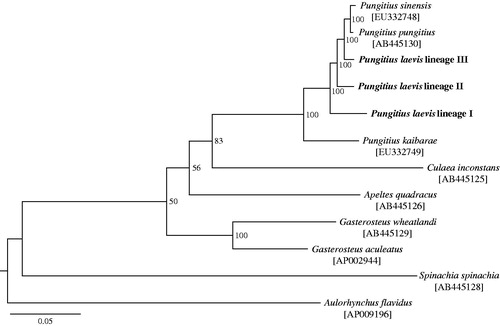
We sequenced genomic DNA of three P. laevis specimens, each belonging to a different lineage (cf. Wang et al. Citation2015; lineage I, 47°25′N, 03°13′E; lineage II, 45°40′N, 00°14′E; lineage III, 47°32′N, 03°38′E). The Illumina HiSeq2000 platform was used with 100 paired-end strategy. Depending on the lineage, between 5.4 and 7.7 million reads were produced and aligned against the P. sinensis mitogenome (Hwang et al. Citation2012a) with bwa-0.5.10 (Li & Durbin Citation2009). In each individual, 100% of the reference genome had onefold coverage; 18.61–99.08% had ≥ 20-fold coverage, with a mean coverage of 14.58–75.02-fold. The complete mitochondrial genomes of P. laevis lineages I, II and III are 16 576, 16 574 and 16 580 bp in length, respectively (GenBank Accession Nos. KT989567-69). Each contained 37 genes (13 protein-coding, 22 transfer RNA (tRNA), two ribosomal RNA (rRNA)) and a control region. The position and direction of these genes were identical to those of other Gasterosteidae mitochondrial genomes (Miya et al. Citation2001; Kawahara et al. Citation2009; Hwang et al. Citation2012a,Citationb). Most of the protein-coding genes, except ND2, COII, ND4 and Cytb, end with a stop codon in all lineages. The overall base composition of the entire genome in each lineage is 27.6–27.7% for A, 26.9–27.0% for T, 17.3% for G and 28.0–28.3% for C. A maximum likelihood tree of 11 Gasterosteidae fishes, including the three P. laevis lineages and the tube-snout (Aulorhynchus flavidus) as an outgroup species, was constructed based on the 37 genes using RAxML v.8.0 (Stamatakis Citation2014) under the GTR + GAMMA model with 37 gene partitions and 100 thorough bootstrap replicates. All Pungitius fish formed a single cluster, with the three P. laevis lineages exhibiting high levels of divergence from each other (). The 37 gene regions (15 584 bp) showed 93.2–98.7% nucleotide identity between the P. laevis lineages and other Pungitius species (i.e. P. pungitius, P. sinensis and P. kaibarae). Nucleotide identity among the three P. laevis lineages was 96.2–97.8%.
Acknowledgements
We thank Pekka Ellonen, Laura Häkkinen, Tiina Hannunen and Sami Karja for help with laboratory and bioinformatic work, and Jacquelin DeFaveri for linguistic check of this manuscript. The sequencing was conducted at Finnish Institute for Molecular Medicine.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by grants (108601 & 118673) from the Academy of Finland.
References
- Gross HP. 1979. Geographic variation in European ninespine sticklebacks, Pungitius pungitius. Copeia. 1979:405–412.
- Hwang DS, Song HB, Lee JS. 2012a. Complete mitochondrial genome of the Chinese stickleback Pungitius sinensis (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:293–294.
- Hwang DS, Song HB, Lee JS. 2012b. Complete mitochondrial genome of the Amur stickleback Pungitius kaibarae (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:313–314.
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M. 2009. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 50:401–404.
- Keivany Y, Nelson JS. 2000. Taxonomic review of the genus Pungitius, ninespine sticklebacks (Gasterosteidae). Cybium. 24:107–122.
- Kottelat M, Freyhof J. 2007. Handbook of European freshwater fishes. Cornol: Publications Kottelat.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenetic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Phylogenet Evol. 18:1993–2009.
- Münzing J. 1969. Variabilität, verbreitung und systematik der arten und unterarten in der gattung Pungitius Coste, 1848 (Pisces, Gasterosteidae). J Zool Syst Evol Res. 7:208–233.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang C, Shikano T, Persat H, Merilä J. (2015). Mitochondrial phylogeography and cryptic divergence in the stickleback genus Pungitius. J Biogegr 42:2334–2348.