Abstract
Ulva prolifera (U. prolifera), a green macroalgae, is widely known as the dominant species of the world’s largest macroalgal blooms in the Yellow Sea, China. In this study, we sequenced and annotated the complete mitochondrial genome of U. prolifera (GenBank accession number: KU161104). The genome consists of circular chromosomes of 61 962 bp and encodes a total of 26 protein-coding genes include nine ribosomal protein genes, five atp genes, three cox genes, eight nad genes and cob gene. Phylogenetic analysis showed U. prolifera clustered into Ulvo phyceae clade and had close genetic relationship with algae Ulva fasciata.
Species of green macroalgae genus Ulva are ubiquitous throughout the world in marine and estuarine habitats, where they show a great ability to acclimate to adverse circumstances and grow rapidly in nutrient rich waters (Tan et al. Citation1999). Ulva prolifera (U. prolifera), a common species in coastal area of China, was identified as the causal species of the world’s largest macroalgal blooms in the Yellow Sea based on morphological characteristics and molecular phylogenetic analysis (Wang et al. Citation2008; Ding et al. Citation2009; Zhao et al. Citation2011, Citation2013). It is also one of the most important seaweeds for producing macroalgal fertilizer, food, biofuel and so on (Hiraoka & Oka Citation2008; Li et al. Citation2010; Zhao et al. Citation2011). Mitogenome of U. prolifera has never been reported before, and here, we determined the complete mitochondrial genome of this seaweed to gain insight into blooming mechanism of the Ulva green tides and genetic diversity of U. prolifera.
The sample of U. prolifera was collected from the coast of Rudong. Healthy algal thalli were transported to our laboratory under low-temperature conditions (4–8 °C) within 48 h. Then, the single intact algae chosen were cleaned of debris and epiphytes, rinsed gently using sterile seawater and cultured in von Stosch's enriched (VSE) culture medium for 2 days. The culture condition was maintained at 24 °C, under a 12:12-h light/dark photoperiod, and light intensity 130–160 μmolm−2s−1. The U. prolifera mitochondrial genome sequence is circular-mapping molecule (GenBank accession number: KU161104). The overall base composition of U. prolifera is T 34.3%, C 17.8%, A 31.8%, G 16.1%, G + C content 33.9%, similar to most of other green algae (30–35%). It contains 26 protein-coding genes, including nine ribosomal protein genes, five atp genes, three cox genes, eight nad genes and cob gene, and 21 tRNA, three rRNA. Majority of genes are encoded on the H-strand except 1 rrl and 2 rrs genes that are encoded on the L-strand. Comparing the gene content of U. prolifera with that of other three Ulvo phyceae algae, the rps4, rps14, rps10, mttB and nad9 are missing in U. prolifera, which implies the occurrence of the horizontal transfer (HGT). All of the 26 protein-coding genes begin from ATG except rps11 which begins with TTA. Three typical complete stop codons were detected in open reading frames of U. prolifera TAA, TGA and TAG. Among these, eight protein-coding genes (rps13, atp9, cob, nad4L, atp1, rps11, nad7, nad2) shared the common termination codon TAG, whereas atp8, rpl14, nad4, nad3, rps19, rpl16, cox1, rps2, atp4 had the common termination codon TAA. And only the termination codon of rps3 was TGA.
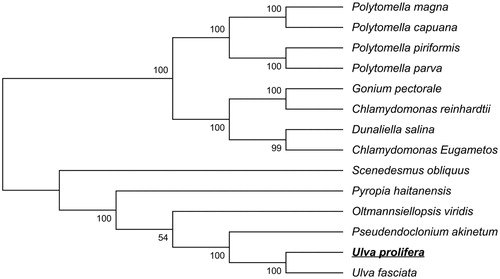
The phylogenetic analysis based on amino acid sequences of seven mt protein-coding genes (cob, cox1, nad1, nad2, nad4, nad5, nad6) from 13 related taxa mitogenome accession number used in this phylogeny analysis: Polytomella magna: KC733827 (Smith et al. Citation2013); Polytomella capuana: NC_010357.1 (Smith et al. Citation2008); Polytomella piriformis: GU108480 + GU108481 (Smith et al. Citation2010); Polytomella parva: NC_016916.1 + NC_016917.1 (Smith et al. Citation2010); Gonium pectorale: AP012493 (Hamaji et al. Citation2013); Chlamydomonas reinhardtii: NC_001638.1 (Vahrenholz et al. Citation1993); Dunaliella salina: NC_012930.1 (Smith et al. Citation2010); Chlamydomonas Eugametos: NC_001872.1 (Denovan-Wright et al. Citation1998); Scenedesmus obliquus: AF204057.1 (Kuck et al. Citation2000); Pyropia haitanensis: NC_017751.1 (Mao et al. Citation2012); Oltmannsiellopsis viridis: NC_008256.1 (Pombert et al. Citation2006); Pseudendoclonium akinetum: NC_005926.1 (Pombert et al. Citation2004); Ulva fasciata: KT364296.1 (Melton & Lopez-Bautista Citation2015). Twelve complete mitogenome entries are under green algae and one from red algae. For the construction of tree, MEGA6 software (Tamura et al. Citation2013) was used with 1000 bootstrap replicates. The phylogenetic tree () showed that the placement of U. prolifera within the Ulvo phyceae.
Figure 1. Phylogenetic tree of U. prolifera with mitochondrial genome sequence. GenBank accession numbers of species used in the tree: Polytomella magna: KC733827; Polytomella capuana: NC_010357.1; Polytomella piriformis: GU108480 + GU108481; Polytomella parva: NC_016916.1 + NC_016917.1; Gonium pectorale: AP012493; Chlamydomonas reinhardtii: NC_001638.1; Dunaliella salina: NC_012930.1; Chlamydomonas Eugametos: NC_001872.1; Scenedesmus obliquus: AF204057.1; Pyropia haitanensis: NC_017751.1; Oltmannsiellopsis viridis: NC_008256.1; Pseudendoclonium akinetum: NC_005926.1; Ulva fasciata: KT364296.1.
We conclude that the complete mtDNA genome sequence obtained in this study would be useful for studying genetic diversity and phylogenetic history of U. prolifera and its related species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by the National Natural Science Foundation of China (Grant No. D8000150127), Ocean Public Welfare Scientific Research Project, China (201205010), Shanghai Plateau Peak Disciplines Project (Discipline name: Marine Science 0707), Training Programme Foundation for Outstanding Young Teachers in Shanghai Universities (ZZZZHY15007) and the Project of Key Laboratory of Integrated Marine Monitoring and Applied Technologies for Harmful Algal Blooms, S.O.A., (MATHAB201502).
References
- Denovan-Wright EM, Nedelcu AM, Lee RW. 1998. Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol Biol. 36:285–295.
- Ding LP, Fei XG, Lu QQ, Deng YY, Lian SX. 2009. The possibility analysis of habitats, origin and reappearance of bloom green alga (Enteromorpha prolifera) on inshore of western Yellow Sea. Chin J Oceanol Limnol. 27:421–424.
- Hamaji T, Smith DR, Noguchi H, Toyoda A, Suzuki M, Kawai-Toyooka H, Fujiyama A, Nishii I, Marriage T, Olson BJSC, et al. 2013. Mitochondrial and plastid genomes of the colonial green alga Gonium pectorale give insights into the origins of organelle DNA architecture within the volvocales. PLoS One. 8:e57177.
- Hiraoka M, Oka N. 2008. Tank cultivation of Ulva prolifera in deep seawater using a new “germling cluster” method. J Appl Phycol. 20:97–102.
- Kuck U, Jekosch K, Holzamer P. 2000. DNA sequence analysis of the complete mitochondrial genome of the green alga Scenedesmus obliquus: evidence for UAG being a leucine and UCA being a non-sense codon. Gene. 253:13–18.
- Li DM, Chen LM, Zhao JS, Zhang XW, Wang QY, Wang HX, Ye NH. 2010. Evaluation of the pyrolytic and kinetic characteristics of Enteromorpha prolifera as a source of renewable bio-fuel from the Yellow Sea of China. Chem Eng Res Des. 88:647–652.
- Mao Y, Zhang B, Kong F, Wang L. 2012. The complete mitochondrial genome of Pyropia haitanensis Chang et Zheng. Mitochondrial DNA. 23:344–346.
- Melton JT III, Lopez-Bautista JM. 2015. De novo assembly of the mitochondrial genome of Ulva fasciata delile (Ulvophyceae, Chlorophyta), a distromatic blade-forming green macroalga. Mitochondrial DNA. Early Online, DOI: 10.3109/19401736.2015.1053071.
- Pombert JF, Beauchamp P, Otis C, Lemieux C, Turmel M. 2006. The complete mitochondrial DNA sequence of the green alga Oltmannsiellopsis viridis: evolutionary trends of the mitochondrial genome in the Ulvophyceae. Curr Genet. 50:137–147.
- Pombert JF, Otis C, Lemieux C, Turmel M. 2004. The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the chlorophyta and suggests a sister-group relationship between the Ulvo phyceae and Chlorophyceae. Mol Biol Evol. 21:922–935.
- Smith DR, Hua J, Archibald JM, Lee RW. 2013. Palindromic genes in the linear mitochondrial genome of the nonphotosynthetic green alga Polytomella magna. Genome Biol Evol. 5:1661–1667.
- Smith DR, Hua J, Lee RW. 2010. Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Curr Genet. 56:427–438.
- Smith DR, Lee RW. 2008. Mitochondrial genome of the colorless green alga Polytomella capuana: a linear molecule with an unprecedented GC content. Mol Biol Evol. 25:487–496.
- Smith DR, Lee RW, Cushman JC, Magnuson JK, Tran D, Polle JE. 2010. The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 10:83.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tan IH, Blomster J, Hansen G, Leskinen E, Maggs CA, Mann DG, Sluiman HJ, Stanhope MJ. 1999. Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha. Mol Biol Evol. 16:1011–1018.
- Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G. 1993. Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Curr Genet. 24:241–247.
- Wang WJ, Wang FJ, Chen SL, Sun XT, Wang XY, Xing SC, Wang J. 2008. PCR amplification and sequence analysis of ITS regions of Enteromorpha prolifera. Mar Fish Res. 29:124–129. (In Chinese with English abstract)
- Zhao J, Jiang P, Liu ZY, Wang JF, Cui YL, Qin S. 2011. Genetic variation of Ulva (Enteromorpha) prolifera (Ulvales, Chlorophyta)-the causative species of the green tides in the Yellow Sea, China. J Appl Phycol. 23:227–233.
- Zhao J, Jiang P, Liu ZY, Wei W, Lin HZ, Li FC, Wang JF, Qin S. 2013. The Yellow Sea green tides were dominated by one species, Ulva (Enteromorpha) prolifera, from 2007 to 2011. Chinese Sci Bull. 58:2298–2302.
- Zhao H, Yan H, Liu M, Zhang CW, Qin S. 2011. Pyrolytic characteristics and kinetics of the marine green tide macroalgae, Enteromorpha prolifera. Chin J Oceanol Limnol. 29:996–1001.
