Abstract
Complete mitochondrial genome of the Peloponnese endemic lizard species Anguis cephallonica is presented in this study. The complete sequence is 17 208 bp long and consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one control region. The gene order is same as in the relative species Anguis fragilis. Length of the 22 tRNA genes varies from 64 bp to 73 bp. The Anguis cephallonica mitogenome base composition is: A (30.5%), T (24.2%), C (30.5%), G (14.8%), with an A + T bias (54.7%). Six protein coding genes have incomplete stop codons. This is the first complete mitogenome described in this species as well as in any endemic Peloponnese lizard. Presented complete mitochondrial genome will form a basis for future comparative analysis within the genus Anguis.
Legless lizards of the genus Anguis Linnaeus, 1758 (slow worms) inhabit a large territory in the Western Palearctic region. Out of five extant species, one is endemic of the Italian Peninsula, two of the Balkans and two other occur in large area of Europe and western Asia (Gvoždík et al. Citation2010, Citation2013). The last species, Peloponnese slow worm, A. cephallonica, has the smallest range and is endemic to the Peloponnese Peninsula, Zakynthos, Cephalonia and Ithaca islands. Originally, A. cephallonica was described as a subspecies of A. fragilis, however, precise studies based on morphometric, allozyme and molecular markers resulted into its recognition as a separate species with intraspecific genetic variability (Grillitsch & Cabela Citation1990; Mayer et al. Citation1991; Gvoždík et al. Citation2010, Thanou et al. Citation2014) and corroborated the Peloponnese Peninsula as an important center of European endemism (cf., Poulakakis et al. Citation2015).
Complete mitochondrial genomes represent a valuable source of information and can be used to infer phylogenetic relationships of various taxa with more precision and detail than short sequences as has been previously successfully demonstrated (Douglas et al. Citation2006; Douglas & Gower Citation2010; Wielstra & Arntzen Citation2011). In order to provide a comprehensive dataset for future phylogenetic studies we decided to sequence complete mitochondrial DNA (mtDNA) of A. cephallonica. We also compared basic genomic characteristics with mitogenome of A. fragilis, the only other slow-worm species sequenced so far (Albert et al. Citation2009).
Tissue sample (muscle from a road-killed individual; voucher number of the Natural History Museum of Crete: NHMC80.3.3.2.) for genetic analysis was collected near Oitylo village (Peloponnese Peninsula, Greece, 36.72086°N; 22.39472°E) (). Total genomic DNA was isolated with Sherlock AX (A&A Biotechnology, Gdynia, Poland) according to the product manual, then amplified in three overlapping fragments and sequenced with primer walking method by Wyzer Biosciences Inc. (Cambridge, USA) and assembled with MITOS WebServer (Bernt et al. Citation2013).
The mitochondrial genome of A. cephallonica (17 208 bp; GenBank KU052866) shares identical organization and gene order with A. fragilis. Overall base composition of A. cephallonica H-strand is as follows: A (30.5%), T (24.2%), C (30.5%), G (14.8%) and slightly differs from A. fragilis: A (29.2%), T (26%), C (28.2%), G (16.6%). Twenty-one out of 38 elements vary in length between both genomes. Control region is shorter by 254 nucleotides in A. cephallonica. The most of the corresponding start codons are the same for protein coding genes (PCGs) excluding ND3 (ATG – A. cephallonica, GTG – A. fragilis). Eleven PCGs have the same start codon – ATG, while COX1 and A. fragilis ND3 require GTG. In A. cephallonica genome, six of PCGs use truncated stop codons (ATP6, COX2, COX3, ND3, ND4, ND5), three genes stop with TAA (ATP8, ND1, ND4L), three with TAG (COX1, CYTB, ND2) and one with AGA (ND6). When compared to A. fragilis genome, stop codons usage differs as the most common stop codons in A. fragilis is TAA (ATP6, ATP8, COX1, ND2, ND4L, ND5), then truncated stop codons (COX2, COX3, ND3, ND4) and TAG (CYTB, ND1) and AGA (ND6).
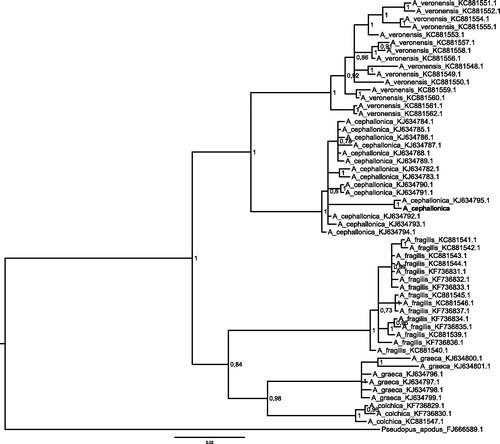
Figure 1. Bayesian phylogenetic tree of five Anguis species ND2 sequence (765 bp) alignment.
Bayesian phylogenetic tree was generated with. MrBayes 3.2.5 (Ronquist et al. Citation2012) using model GTR + I +G as suggested by jModelTest 2.1.7 (Guindon & Gascuel Citation2003; Darriba et al. Citation2012). 10 000 000 MCMC repetitions with burn-in of 25% was used. ND2 sequence of Anguis cephallonica from the present study is bolded and grouped together with other A. cephallonica representatives. Tree was rooted with Pseudopus apodus ND2 sequence and all GenBank accession numbers of all used sequences are present on the tree. Bayesian posterior probabilities are showed with nodes.
Acknowledgements
D. Jab., P.M. & D. Jan. were supported by the grants VEGA 1/0073/14, UK/20/2014 and UK/37/2015. We thank the Greek Ministry of Agricultural Development and Foods for the permit No. 92221/2067. This work was supported by the Wroclaw University of Environmental Sciences, Faculty of Biology and Animal Science under Grant for Youth Scientists. The publication was supported by Wroclaw Centre of Biotechnology, programme “The Leading National Research Centre (KNOW) of Wroclaw University of Environmental and Life Sciences for years 2014–2018”.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Albert EM, San Mauro D, García-París M, Rüberb L, Zardoya R. 2009. Effect of taxon sampling on recovering the phylogeny of squamate reptiles based on complete mitochondrial genome and nuclear gene sequence data. Gene 441:12–21.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Douglas DA, Gower DJ. 2010. Snake mitochondrial genomes: phylogenetic relationships and implications of extended taxon sampling for interpretations of mitogenomic evolution. BMC Genomics 11:14.
- Douglas DA, Janke A, Arnason U. 2006. A mitogenomic study on the phylogenetic position of snakes. Zool Scr. 35:545–558.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Grillitsch H, Cabela A. 1990. Zum Systematischen Status der Blindschleichen (Squamata: Anguidae) der Peloponnes und der südlichen Ionischen Inseln (Griechenland). Herpetozoa 2:131–153.
- Gvoždík V, Benkovský N, Crottini A, Bellati A, Moravec J, Romano A, Sacchi R, Jandzik D. 2013. An ancient lineage of slow worms, genus Anguis (Squamata: Anguidae), survived in the Italian Peninsula. Mol Phylogenet Evol. 69:1077–1092.
- Gvoždík V, Jandzik D, Lymberakis P, Jablonski D, Moravec J. 2010. Slow worm, Anguis fragilis (Reptilia: Anguidae) as a species complex: genetic structure reveals deep divergences. Mol Phylogenet Evol. 55:460–472.
- Mayer W, Grillitsch H, Cabela A. 1991. Protein elektrophoretische Untersuchungen zur Systematic der südgriechischen Blindschleichen (Squamata: Anguidae: Anguis). Herpetozoa 4:157–165.
- Poulakakis N, Kapli P, Lymberakis P, Trichas A, Vardinoyiannis K, Sfenthourakis S, Mylonas M. 2015. A review of phylogeographic analyses of animal taxa from the Aegean and surrounding regions. J Zoolog Syst Evol Res. 53:18–32.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Thanou E, Giokas S, Kornilios P. 2014. Phylogeography and genetic structure of the slow worms Anguis cephallonica and Anguis graeca (Squamata: Anguidae) from the southern Balkan Peninsula. Amphibia-Reptilia 35:263–269.
- Wielstra B, Arntzen JW. 2011. Unraveling the rapid radiation of crested newts (Triturus cristatus superspecies) using complete mitogenomic sequences. BMC Evol Biol. 11:162.
