Abstract
In this study, the complete mitochondrial genome of Volutharpa perryi has been determined for the first time. The mitogenome is of 15 255 bp in length, including 13 protein-coding genes, two ribosomal RNA genes and 22 transfer RNA genes. The overall base composition is 29.6%, 38.6%, 15.6% and 16.2% for A, T, C and G, respectively. The 13 PCGs of V. perryi and other 12 mollusk species were used for phylogenetic analysis by maximum-likelihood method, and the phylogenetic tree demonstrated the close relationship of V. perryi to species of Buccinoidea. The results are expected to provide useful molecular data for species identification and further phylogenetic studies of genus Volutharpa.
Volutharpa perryi, a kind of marine gastropod mollusks in genus Volutharpa of family Buccinidae, inhabiting shallow soft mud or mud sandy seabeds, is mainly distributed in Japan, Korea and Northern Yellow Sea of China (Zhang Citation2008). Many researches had described V. perryi as fishery resource (Ganmanee et al. Citation2004; Zhang et al. Citation2015), but few reports about this species at the molecular level were available (Hou et al. Citation2013). In this study, we determined the complete mitochondrial genome of V. perryi for the first time, expecting to contribute to the better understanding of this species and provide information for the taxonomic and phylogenetic study of the genus Volutharpa.
The specimen of V. perryi were collected from Zhangzi Island of China in June 2015. Standard phenol-chloroform method (Sambrook & Russel Citation2002) was used to extract total genomic DNA from the muscle tissue. Based on the sequences of COX1, 12S rRNA and 16S rRNA (GenBank accession number HQ834060.1, HQ833869.1 and HQ833930.1, respectively), six pairs of primers were designed: three pairs were employed to confirm the known sequences and the other three pairs to obtain three gaps. Long and accurate polymerase chain reaction method was used to determine the three unknown fragments: ∼3600 bp from COX1 to 12S rRNA, ∼1000 bp from 12S to 16S rRNA and ∼9500 bp from 16S rRNA to COX1. After sequenced and assembled, the complete mitogenome sequence was got and deposited into GenBank with the accession number KT382829.
The complete mitogenome of V. perryi is a closed circular molecule of 15 255 bp, consists of 13 protein-coding genes, two ribosomal RNA genes and 22 transfer RNA genes. The total base composition is 29.6% for A, 38.6% for T, 15.6% for C, 16.2% for G, with the AT content (68.2%) higher than that of GC. All genes are encoded on H-strand except seven tRNA genes. All of the 13 PCGs use ATG as the start codon, nine of them use TAA as the stop codon while ND4L, ND4, ND3 and ND2 end with TAG. The 12S rRNA (881bp) and 16S rRNA (1357 bp) is adjacent in position except the tRNA-Val located between them. The tRNA genes were identified by ARWEN online service (http://130.235.46.10/ARWEN/), all of them could be folded in the typical cloverleaf structure except tRNA-Ser.
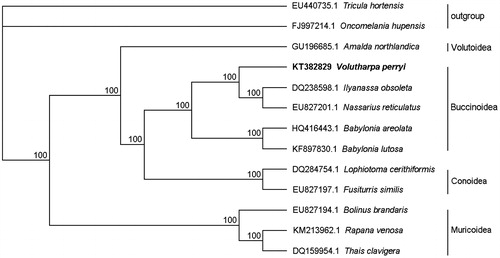
To understand the phylogenetic relationships, the amino acid sequence of 13 PCGs of V. perryi and other 12 mollusks were concatenated for analysis. Tree constructed by Phylip-3.695 using maximum-likelihood method was shown (), and the formation of major clades was identical to the conclusion made from the morphological taxonomy. Some taxonomic studies had reported that V. perryi was allocated to genus Buccinum and named as Buccinum perryi (Zhang & Qi Citation1962; Zhao et al. Citation1982). However, no information about the complete mitogenome of Buccinum was found in GenBank, there is a lack of comparison of Buccinum with V. perryi. In the phylogenetic tree, V. perryi showed a closer relationship with the family Nassariidae, so it is necessary to combine morphology, molecular biology and information of relative species for the taxonomy of V. perryi in further study.
Funding information
The work was supported by Project of National Infrastructure of Fishery Germplasm Resource (2012DKA30470-012) and Liaoning Scientific Research Program of Ocean and Fisheries Department (No. 201519).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ganmanee M, Narita T, Sekiguchi H. 2004. Long-term investigation of spatio-temporal variations in faunal composition and species richness of megabenthos in Ise Bay, central Japan. J Oceanogr. 60:1071–1083.
- Hou L, Dahms HU, Dong CY, Chen YF, Hou HC, Yang WX, Zou XY. 2013. Phylogenetic positions of some genera and species of the family Buccinidae (Gastropoda: Mollusca) from China based on ribosomal RNA and COI sequences. Chinese Sci Bull. 58:2315–2322.
- Sambrook J, Russell DW. 2002. Molecular cloning: a laboratory manual, 3rd ed. Beijing, China: Science Press.
- Zhang JL, Xiao N, Zhang SP, Xu FS, Zhang SQ. 2015. A comparative study on the macrobenthic community over a half century in the Yellow Sea, China. J Oceanogr. [Epub ahead of print]. DOI: 10.1007/s10872-015-0319-z.
- Zhang SP. 2008. Atlas of marine mollusks in China. Beijing, China: Ocean Press.
- Zhang X, Qi ZY. 1962. Economic animals fauna of China: marine mollusca. Beijing, China: Science Press.
- Zhao RY, Cheng JM, Zhao DD. 1982. Mollusca graphy around the Dalian Coast. Beijing, China: Ocean Press.