Abstract
In this study, the complete mitogenome of Buccinum pemphigum has been determined. The complete mitochondrial genome is 15 265 bp in length, including 13 protein-coding genes, two rRNA genes and 22 tRNA genes. The total base composition is 30.1% A, 15.8% G, 15.1% C and 39.0%T, with a high AT content of 69.1%. Tree constructed using maximum-likelihood (ML) phylogenetic method, demonstrated that B. pemphigum has a close relationship to Volutharpa perryi clustered in Buccinoidea. First, this complete mitogenome of Buccinum will facilitate the development of new DNA markers for species identification.
Buccinum pemphigum, belonging to genus Buccinum of family Buccinidae, is a common species distributing in the northern Yellow Sea, the Bering Sea and Japan (Zhao et al. Citation1982). They are gregatious living in the subtidal zone and shallow sea. In this study, we determined the complete mitogenome sequence of B. pemphigum, contributing to species identification and further phylogenetic studies of Buccinidae.
The sample of B. pemphigum was collected from Zhangzi island, Liaoning Province, China, in September, 2014. Whole-genomic DNA was extracted from muscle tissue on the basis of the standard phenol/chloroform method (Sambrook et al. Citation2002). According to the sequence of 16S ribosomal RNA gene (GenBank accession number JN052934.1) and the partial sequence of COX1 (GenBank accession number EU883628.1), two pairs of primers were designed to obtain two large fragments through the method of long and accurate polymerase chain reaction (LA-PCR): a 5334 bp fragment from COX1 gene to 16S ribosomal RNA gene and a 9545 bp fragment from 16S rRNA gene to COX1 gene. After assembling and alignment, we deposited the complete mitogenome sequence of B. pemphigum to GenBank database with accession number KT962044. Transfer RNA genes analysis was performed by ARWEN v1.2 (Laslett & Canback Citation2008).
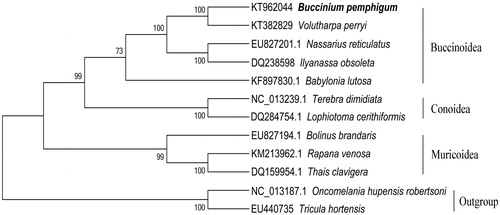
The complete mitogenome of B. pemphigum is a closed circular molecule of 15 265 bp in length, containing a set of 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes and 22 transfer RNA (tRNA) genes, and the base composition is 30.1% A, 39.0% T, 15.1% C and 15.8% G, with an AT content of 69.1%. Its organization and gene order are similar to those of other snail mitochondrial genomes (Feldmeyer et al. Citation2010; Groenenberg et al. Citation2012). All PCGs are initiated with a typical ATN start codon (ATT, ATC, ATG and ATA) except for ND4 with GTG. TAA is the stop codon for most PCGs and ND3, ND4L and ND5 genes end with TAG. Most mitochondrial genes of B. pemphigum, including 13 PCGs, are encoded on the H-strand, except for eight tRNA genes, which are encoded on the L-strand. There are 21 non-coding regions varying from 1 bp to 76 bp in length and four gene overlaps (total 7 bp). Maximum-likelihood (ML) tree was constructed by MEGA 5.0 (Tamura et al. Citation2011) using 13 PCGs from 12 species published in NCBI including Volutharpa perryi, Ilyanassa obsoleta, Nassarius reticulatus and so on. Tricula hortensis and Oncomelania hupensis robertsoni were chosen as outgroup to confirm the phylogenetic relationship. The result showed that B. pemphigum and V. perryi were clustered in one clade belonging to Buccinidae (). The former conclusion made for morphological taxonomy was identificated. We hoped that the study contributes to species identification and may facilitate further investigation of the molecular evolution of Neogastropoda.
Figure 1. Maximum-likelihood phylogenetic analysis for Buccinum pemphigum based on the concatenated amino acids sequences of 13 mitochondrial protein-coding genes from Ilyanassa obsolete, Nassarius reticulates and other nine species. The numbers on each node indicate the bootstrap value. The number at the left of species name is GenBank accession number.
Acknowledgements
We would like to thank Professor Feng Zhang for his instructions in this study.
Disclosure statement
The authors alone are responsible for the content and writing of the paper.
Funding information
The work was supported by Liaoning Scientific Research Program of Ocean and Fisheries Department (No. 201519).
References
- Feldmeyer B, Hoffmeier K, Pfenninger M. 2010. The complete mitochondrial genome of Radix balthica (Pulmonata, Basommatophora), obtained by low coverage shot gun next generation sequencing. Mol Phylogenet Evol. 57:1329–1333.
- Groenenberg DS, Pirovano W, Gittenberger E, Schilthuizen M. 2012. The complete mitogenome of Cylindrus obtusus (Helicidae, Ariantinae) using Illumina next generation sequencing. BMC Genomics. 13:114.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Sambrook J, Russell DW. 2002. Molecular cloning: a laboratory manual, 3rd ed. Beijing, China: Science Press.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Zhao RY, Cheng JM, Zhao DD. 1982. Dalian marine mollusks. Beijing, China: Ocean Press (in Chinese).
