Abstract
The complete mitochondrial genome of a frog species Oreolalax major is determined. This mitogenome length is 17 431 bp, containing 13 protein-coding genes, two rRNA genes, 23 tRNA genes and a control region (D-loop). Compared with most other vertebrates, this mitogenome appears a tandem duplication of tRNAMet gene. The tRNATrp gene of Oreolalax major translocates from the “WANCY” tRNA cluster to upstream of D-loop. As the first report of the mitogenome sequence from the genus Oreolalax, it will provide fundamental data for further research of phylogeny and biogeography with this genus.
Keywords:
The genus Oreolalax (Myers & Leviton Citation1962) belongs to Leptobrachiinae (Delorme et al. Citation2006), Megophryidae. Except one species is in Vietnam (Nguyen et al. Citation2013), the other 17 species are all in southwest of China. Oreolalax major is one of them. The phylogenetic relationships within this genus are still in doubt. It is necessary to study mitochondrial genome on species of Oreolalax, exploring their phylogenetic relationship and geographical distribution pattern by researching on mitochondrial gene arrangement and base composition of genes. The specimen of O. major was collected from Luding County, Sichuan Province, China (Voucher no. CIB-LX548). In this study, we designed 13 pairs of specific primers. Standard PCR and LA-PCR methods were used to amplify the mitochondrial genome of this species. We determined the complete mitogenome of O. major, the first mitogenome from the genus Oreolalax. The GenBank accession number is KU127230.
The gene order and structure of O. major mitogenome was approximately similar to most other amphibians (Chen et al. Citation2011; Xiang et al. Citation2013a,Citationb). The length of this mitogenome was 17 431 bp, containing 13 protein-coding genes, two rRNA genes, 23 tRNA genes and a control region (D-loop). The base composition of the whole genome was 28.8% A, 32.4% T, 24.5% C and 14.4% G. Except for ND6 gene and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser (UCN), Glu and Pro) encoded on the L-strand, the remaining genes were encoded on the heavy strand (H-strand). The 23 tRNA genes with the size ranging from 64 to 75 bp were interspersed along the whole genome. As observed in Vibrissaphora boringii of genus Vibrissaphora (Xu et al. Citation2014), this mitogenome had also two tRNAMet genes (tRNAMet1 and tRNAMet2 genes) derived from a tandem duplication. The tRNATrp gene translocated from the cluster of WANCY to the upstream of Cytb gene. The putative origin replication of L-strand replication (OL) located between tRNAAsn gene and tRNACys gene. In O. major, nine protein-coding genes started with the common initiation codon ATG, except for ND3 with ATT. In addition, COI, ND5 and ND6 started with GTG. Stop codons were variable for all protein-coding genes. Six genes (ND1, ND2, COII, ATP8, ND3 and ND4L) used complete stop codon TAR (TAA and TAG), whereas three genes (COI, ND5 and ND6) ended with AGG. Four genes (ATP6, COIII, ND4 and Cytb) ended with incomplete stop codon with T, which may be presumably completed by posttranscriptional polyadenylation with poly A tail (Ojala et al. Citation1981). The length of 12S rRNA and 16S rRNA were 944 bp and 1596 bp, respectively. They located between tRNAPhe and tRNALeu (UUR) genes, separated by the tRNAVal gene. A relatively large spacer region appeared between tRNAGln and tRNAMet1 genes up to 141 bp. There was also 41 bp spacer region between two tRNAMet genes. The D-loop region located between tRNATrp and tRNAPhe genes. The length of this region was 1664 bp with high A + T of 63.9%.
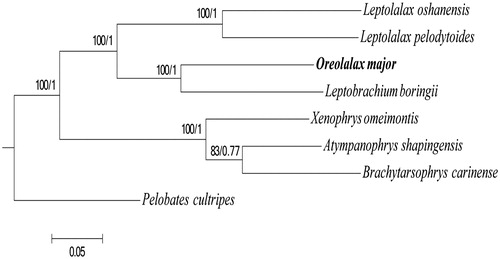
Based on the reported data of seven species of Mesobatrachia, 12 mitochondrial protein-coding genes (PCGs) were used to construct related phylogenetic trees. Maximum-likelihood (ML) and Bayesian inference (BI) methods resulted in the same tree topology. The phylograms are shown in .
Figure 1. The phylogenetic tree inferred by Phyml_3.0 (Guindon et al. Citation2010) and MrBayes v3.2.1 (Ronquist et al. Citation2012). The digit along the branches show support values (bootstrap value/posterior probability). The GenBank accession numbers we used are as follows: Leptolalax oshanensis (KC460337), Leptolalax pelodytoides (JX564874), Leptobrachium boringii (KJ630505), Xenophrys omeimontis (KP728257), Atympanophrys shapingensis (JX458090), Brachytarsophrys carinense (JX564854), Pelobates cultripes (NC_008144).
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper. This study was supported by the National Natural Sciences Foundation of China (NSFC – 31372174 granted to Feng Xie, NSFC – 31201702 granted to Bin Wang and NSFC – 31172055 granted to Cheng Li).
References
- Chen GY, Wang B, Liu JY, Xie F, Jiang JP. 2011. Complete mitochondrial genome of Nanorana pleskei (Amphibia: Anura: Dicroglossidae) and evolutionary characteristics of the amphibian mitochondrial genomes. Curr Zool. 57:785–805.
- Delorme M, Dubois A, Grosjean S. 2006. Une nouvelle ergotaxinomie des Megophryidae (Amphibia, Anura). Alytes. 24:6–21.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Myers GS, Leviton AE. 1962. Generic classification of the high-altitude pelobatid toads of Asia (Scutiger, Aelurophryne, and Oreolalax). Copeia. 1962:287–291.
- Nguyen TQ, Phung TM, Minh DL, Ziegler T, Böhme W. 2013. First record of the genus Oreolalax (Anura: Megophryidae) from Vietnam with description of a new species. BioOne. 2013:213–222.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Xiang TM, Wang B, Jiang JP, Li C, Xie F. 2013a. The complete mitochondrial genome of Megophrys shapingensis (Amphibia, Anura, Megophryidae). Mitochondrial DNA 24:43–45.
- Xiang TM, Wang B, Liang XX, Jiang JP, Li C, Xie F. 2013b. The complete mitochondrial genome of Paramegophrys oshanensis (Amphibia, Anura, Megophryidae). Mitochondrial DNA. 24:472–474.
- Xu QP, Liu SL, Wan RZ, Yue BS, Zhang XY. 2014. The complete mitochondrial genome of the Vibrissaphora boringii (Anura: Megophryidae). Mitochondrial DNA. 24:472–474.
