Abstract
We determined the complete mitochondrial genome of Metapenaeopsis dalei (Rathbun, 1902), which is collected from East China Sea (124°E and 33.5°N). Total mitochondrial genome length of M. dalei was 15 939 bp, in which 13 proteins, two ribosomal RNAs, 22 transfer RNAs and a putative control region were encoded. The gene arrangement of M. dalei was well conserved with nine known Penaeidae mitochondrial genomes from COX1 to tRNATyr. The protein-coding genes started with ATN except for COX1 in which ACG is used. Four genes (COX2, COX3, ND3 and ND5) exhibited an incomplete stop codon. Nucleotide sequence identity of M. dalei mitochondrial genome to those of nine Penaeidae species ranged from 78% to 80%. Based on the COI region, M. dalei is most closely related to Penaeus notialis.
The Kishi velvet shrimp, Metapenaeopsis dalei (Decapoda: Penaeidae) is distributed along the coastal and brackish waters in North China, Korea and Japan (Holthuis Citation1980; Cha et al. Citation2001). As M. dalei is one of the commercially important shrimp species in southern and western coastal waters in Korea, its basic biology and life history have been documented (Choi & Hong Citation2001a,Citationb; Choi et al. Citation2003, Citation2005). Metapenaeopsis dalei belongs to the genus Metapenaeopsis, which includes the most diverse species among Penaeidae prawn. Although its morphological characteristics has been reported such as the asymmetrical petasma in males (Dall et al. Citation1990), relatively little information on the molecular phylogeny has been published (Tong et al. Citation2000; Cheng et al. Citation2015). To our best knowledge, no mitochondrial genome sequences belonging to genus Metapenaeopsis is currently listed in GenBank database. Here we provide the first report about full mitochondrial genome sequence of genus Metapenaeopsis. The M. dalei examined in this study was collected in the East China Sea (124°E and 33.5°N) during the regular survey by National Fisheries Research and Development Institute (NFRDI) in Korea.
Total mitochondrial genome of M. dalei was here determined by the contig assembly obtained by next generation sequencing (NGS) using MiSeq platform (Illumina, San Deigo, IL) and ambiguous sequences were reconfirmed by the typical Sanger sequencing strategy of the cloned PCR products. A 15 939 bp of full length mitochondrial genome (GenBank accession no. KU050082) encoded 13 proteins, two ribosomal RNAs and 22 transfer RNAs, as well as a putative control region. The gene arrangement of all known mitochondrial genomes of Penaeidae including M. dalei was well conserved from COX1 to tRNATyr. The protein-coding genes start with ATN except for COX1 in which ACG is used. Four genes (COX2, COX3, ND3 and ND5) exhibit an incomplete stop codon (T–). The length of LrRNA and SrRNA gene was 1355 bp and 850 bp, respectively. The tRNA genes were spread over the entire genome and were located on both strands. Twenty of them were identified using tRNAscan-SE 1.21 (Lowe & Eddy Citation1997) and the other variable two tRNA genes (tRNAAsn and tRNASer(4672∼4738)) were confirmed by the comparison of mitochondrial genomes of previously known nine Penaeidae prawns (). The mitochondrial genome encoded 22 tRNA genes ranging in size from 65 to 72 nucleotides. The putative control region (996 bp) is the major non-coding region with a high A + T content in M. dalei (80.8%), which is the most variable region within the total mitochondrial genome. Nucleotide sequence of M. dalei mitochondrial genome shared 78–80% identity to those of nine Penaeidae species, showing the highest identity to Metapenaeus ensis. When only COI region was compared, M. dalei is most closely related to Penaeus notialis (Pérez-Farfante, 1967), which is caught in eastern Atlantic and west African coast.
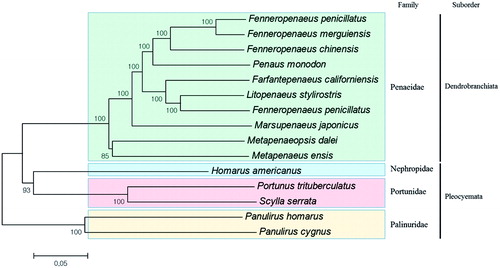
Figure 1. Phylogenetic tree of mitochondrial genomes from six species belonging to Palinuridae with other decapod crustaceans. Phylogenetic tree was constructed using molecular evolutionary genetics analysis ver. 6.0 (MEGA6, MEGA Inc., Englewood, NJ) program with the minimum evolution algorithm. The evolutionary distances were computed using the Kimura 2-parameter method. Total mitochondrial genome sequences used for the analysis include 10 from Penaeidae (Fenneropenaeus penicillatus; NC_026885, Fenneropenaeus merguiensis; NC_026884, Metapenaeus ensis; NC_026834, Marsupenaeus japonicus; NC_007010, Litopenaeus stylirostris; NC_012060, Litopenaeus vannamei; NC_009626, Fenneropenaeus chinensis; NC_009679, Farfantepenaeus californiensis; NC_012738, Penaeus monodon; NC_02184 and Metapenaeopsis dalei; KU050082), two from Portunidae (Portunus trituberculatus; NC_005037, Scylla serrata; HM590866), two from Palinuridae (Panulirus homarus; NC_016015, Panulirus Cygnus; NC_028024) and one from Nephropidae (Homarus americanus; NC_015607).
Disclosure statement
This work was supported by a grant from the National Fisheries Research and Development Institute (R2015024).
References
- Cha H, Lee J, Park C, Baik C, Hong S, Park J, Lee D, Choi Y, Hwang K, Kim Z. 2001. Shrimps of the Korean waters. Busan: National Fisheries Research and Development Institute. p. 1–188.
- Cheng J, Sha Z-l, Liu R-y. 2015. DNA barcoding of genus Metapenaeopsis (Decapoda: Penaeidae) and molecular phylogeny inferred from mitochondrial and nuclear DNA sequences. Biochem Syst Ecol. 61:376–384.
- Choi J, Hong S. 2001a. Larval development of the kishi velvet shrimp, Metapenaeopsis dalei (Rathbun)(Decapoda: Penaeidae), reared in the laboratory. Fish Bull. 99:275–291.
- Choi J, Kim J, Kim J, Cha H, Hong S. 2003. Gonad development and genital organs of Metapenaeopsis dalei (Crustacea: Decapoda: Penaeidae) in the Korean waters. J Korean Fish Soc. 36:619–624.
- Choi JH, Hong SY. 2001b. Molt-staging and setal morphology of Metapenaeopsis dalei (Decapoda: Penaeidae). Korean J Fish Aquatic Sci. 34:38–42.
- Choi JH, Kim JN, Ma CW, Cha HK. 2005. Growth and reproduction of the kishi velvet shrimp, Metapenaeopsis dalei (Rathbun, 1902) (Decapoda, Penaeidae) in the western sea of Korea. Crustaceana. 78:947–963.
- Dall W, Hill B, Rothlisberg P, Sharples D. 1990. The biology of the Penaeidae. Adv Mar Biol. 27:489.
- Holthuis LB, 1980. FAO species catalogue. Volume 1 – Shrimps and prawns of the world. An annotated catalogue of species of interest to fisheries. Rome: Food and Agriculture Organization of the United Nations.
- Lowe TM, Eddy SR, 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 25:955–964.
- Pérez Farfante I. 1967. A new species and two new subspecies of shrimp of the genus Penaeus from the Western Atlantic. Proceedings of the Biological Society of Washington. 80:83–100.
- Tong J, Chan T, Chu K. 2000. A preliminary phylogenetic analysis of Metapenaeopsis (Decapoda: Penaeidae) based on mitochondrial DNA sequences of selected species from the Indo-West Pacific. J Crust Biol. 20:541–549.
