Abstract
In this study, the total mitochondrial genome sequence of Andrias davidianus (Caudata: Cryptobrachidae) was determined. The genome is 16,519 bases in length. It consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and 2 non-coding regions. These results will contribute to the natural resources conservation and species identification Of A. davidianus.
The Chinese giant salamanders (Andrias davidianus), belonging to the Order Caudata and the Family Cryptobranchidae, is one of the world’s largest amphibian species (Dong et al. Citation2011). Andrias davidianus may represent the transition steps from aquatic to terrestrial life in the evolution history of vertebrate system (Wang et al. Citation2013), it has an important value in scientific research of phylogeny relationship of land vertebrate evolution.
Because of its maternal mode of inheritance and relative lack of recombination (Saccone et al. Citation1999; Arnason et al. Citation2002). The complete mitochondrial genome sequence is a useful tool for the analysis of genetic diversity of endangered species (Chen et al. Citation2012; Min & Park Citation2009; Peng et al. Citation2006).
In this study, an individual A. davidianus was sampled in Shaoxing, Zhejiang province, China (29.77°N, 120.58°E) and was collected in the Animal specimen room of Shaoxing University (No. 405). The complete mitochondrial genome sequence of the sample was sequenced and characterized. The PCR amplification was performed using total DNA as template, and the parameters are initial denaturation at 95 °C for 4 min, followed by 35 cycles of amplification (94 °C for 45 s, 52 °C or 58 °C for 30 s and 72 °C for 90 s), and one final cycle of 10 min at 72 °C.
The complete mitochondrial genome was determined to be 16,519 bp in length (GenBank accession no. KT119359). It consists of 22 tRNA genes, the large and small rRNA unit genes, 13 protein-coding genes (PCGs), and 2 non-coding regions: origin of light-strand replication (OL) and control region (CR).
Except for ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro), all of the other mitochondrial genes are encoded on the heavy strand (H-strand). Total base composition of the mitochondrial genome of A. davidianus: 31.91% A, 32.81% T, 20.97% C, and 14.31% G, respectively.
Except for COX I with a GTG start codon, the remaining protein-coding genes initiate with ATG. Six protein-coding genes end with TAA (COX I, ATP8, ATP6, ND3, ND4L, and ND6). Incomplete stop codons are found in ND5, ND4(TA-), as well as ND1, ND2, COX II, COX III, and Cyt b (T–), respectively.
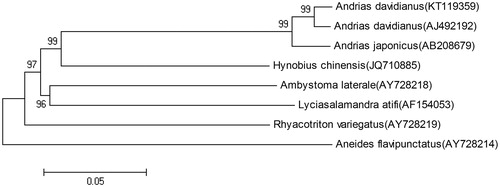
A phylogenetic tree was constructed based on the complete mitochondrial genome sequences of A. davidianus and other salamanders using the neighbour-joining method (). The results showed that A. davidianus and A. japonicus are clustered together, and showed close relationship with other salamanders including Hynobius chinensis (Tang et al. Citation2015) and Lyciasalamandra atifi, while it showed distant kinship with other salamanders.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Arnason U, Adegoke JA, Bodin K, Born EW, Esa YB, Gullberg A, Nilsson M, Short RV, Xu X, Janke A. 2002. Mammalian mitogenomic relationships and the root of the eutherian tree. Proc Natl Acad Sci USA. 99:8151–8156.
- Chen DX, Chu WY, Liu XL, Nong XX, Li YL, Du SJ, Zhang JS. 2012. Phylogenetic studies of three sinipercid fishes (Perciformes: Sinipercidae) based on complete mitochondrial DNA sequences. Mitochondrial DNA. 23:70–76.
- Dong W, Zhang X, Yang C, An J, Qin J, Song F, Zeng W. 2011. Iridovirus infection in Chinese giant salamanders, China, 2010. Emerging Infect Dis. 17:2388–2389.
- Min GS, Park JK. 2009. Eurotatorian paraphyly: revisiting phylogenetic relationships based on the complete mitochondrial genome sequence of Rotaria rotatoria (Bdelloidea: Rotifera: Syndermata). BMC Genomics. 10:533.
- Peng ZG, Wang J, He SP. 2006. The complete mitochondrial genome of the helmet catfish Cranoglanis bouderius (Siluriformes: Cranoglanididae) and the phylogeny of otophysan fishes. Gene. 376:290–297.
- Saccone C, Giorgi CD, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209.
- Tang D, Xu T, Sun Y. 2015. Hynobiidae origin in middle Cretaceous corroborated by the new mitochondrial genome of Hynobius chinensis. Mar Genomics. 22:37–44.
- Wang L, Yang H, Li F, Zhang Y, Yang Z, Li Y, Liu X. 2013 . Molecular characterization, tissue distribution and functional analysis of macrophage migration inhibitory factor protein (MIF) in Chinese giant salamanders Andrias davidianus. Dev Comp Immunol. 39:161–168.