Abstract
Eastern paradise fish, Polynemus dubius (also called P. longipectoralis), lives in rivers of the Southeastern Asia. Although its whole genome is not available, we first obtained its complete mitochondrial genome through next-generation sequencing. The length of its mitochondrial genome is 16 568 bp with 13 protein-coding genes, two rRNA genes, 22 tRNA genes and one control region. The distribution and arrangement of genes in the mitochondrial genome are similar to those of other vertebrates. The GC content is 43.08%, similar to the reported P. paradiseus. A phylogenetic tree was constructed to compare with nine related species by using MEGA 6.0. Through phylogenetic analysis, the relationship of species within the Polynemus genus is determined.
Eastern paradise fish, Polynemus dubius (Motomura Citation2003), also called Polynemus longipectoralis, lives in the Kangsar and Muar rivers (western Malaysia in Malay Peninsula), Musi and Batanghari rivers (southeastern Sumatra, Indonesia), and Sampit and Barito rivers (southern Kalimantan, Indonesia) (Motomura & Tsukawaki Citation2006). We obtained its samples from the wild (Guangdong Province, China) and stored at China National Genebank (accession no. GZ2014122001). Although its genome is not available, we sequenced its mitochondrial genome using Illumina Hiseq 4000 sequencing platform (Illumina Inc., San Diego, CA) (Tang et al. Citation2015). Finally, we assembled the complete mitochondrial genome. By utilizing the acquired mitochondrial genome, we can easily distinguish and even protect this precious fish.
Total length of the complete mitochondrial genome of P. dubius (accession no. KU199001) is 16 555 bp (which contains 13 protein-coding genes while nad6 in a reverse orientation), two rRNA genes (12s rRNA with 942 bp and 16s rRNA with 1704 bp in length), 22 tRNA genes and one control region (774 bp). The length of the 22 tRNA genes ranges from 64 bp to 73 bp. However, 10 of the 22 tRNA genes are located in a reverse orientation (Wyman et al. Citation2004). The start codon of 11 protein-coding genes is ATG, while the rest two genes are starting with GTG and GAC, respectively. Overall, the composition of complete sequence is A (29.91%), T (27.01%), C (27.93%), G (15.15%), and the overall GC-content is 43.08%.
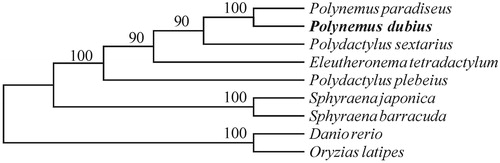
To evaluate accuracy of the assembly and annotation, we downloaded complete mitochondrial genomes of nine reported fish species, including Eleutheronema tetradactylum, Polydactylus plebeius, Polydactylus sextarius, Polynemus paradiseus, Sphyraena barracuda, Sphyraena japonica, Danio rerio and Oryzias latipes for comparison. We used Muscle Program (vision: v3.8.31; Edgar Citation2004) to execute the whole genome alignment. Subsequently, we chose the conserved blocks by the online Gblocks (http://phylogeny.lirmm.fr/). A neighbour-joining tree based on the maximum-likelihood method was constructed based on the conserved blocks by using MEGA 6.0 Program (Tamura et al. Citation2013). Our results demonstrate that the P. dubius is much closer to P. paradiseus than P. sextarius ().
Figure 1. The rectangular phylogenetic tree based on the conserved blocks within mitochondrial genomes of ten related species. The analysis was performed using the MEGA 6.0 software. Accession number: Eleutheronema tetradactylum (NC_021620.1), Polydactylus plebeius (NC_026235.1), Polydactylus sextarius (NC_027088.1), Polynemus paradiseus (NC_026236.1), Sphyraena barracuda (NC_022484.1), Sphyraena japonica (NC_022489.1), Danio rerio (NC_002333.2) and Oryzias latipes (NC_004387.1)
Funding information
The work was supported by Special Project on the Integration of Industry, Education and Research of Guangdong Province (No. 2013B090800017), Shenzhen Special Program for Future Industrial Development (No. JSGG20141020113728803), Special Fund For State Oceanic Administration Scientific Research in the Public Interest (No. 201305018) and Shenzhen Scientific R & D Grant (No. CXB201108250095A).
Disclosure statement
The authors have declared no conflict of interests. We are responsible for the content and writing of the paper.
References
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Motomura H. 2003. A new species of freshwater threadfin, Polynemus aquilonaris, from Indochina, and redescription of Polynemus dubius Bleeker, 1853 (Perciformes: Polynemidae). Ichthyological Res. 50:154–163.
- Motomura H, Tsukawaki S. 2006. New species of the threadfin genus Polynemus (Teleostei: Polynemidae) from the Mekong River basin, Vietnam, with comments on the Mekong species of Polynemus. Raffles Bull Zool. 54:459–464.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tang M, Hardman CJ, Ji Y, Meng G, Liu S, Tan M, Yang S, Moss ED, Wang J, Yang C. 2015. High‐throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods Ecol Evol. 6:1034–1043.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
