Abstract
We sequenced the complete mitochondrial genome from Minla ignotincta. The genome sequence was 17 868 bp in length, and the gene arrangement and contents were identical to those of previously reported Timaliidae mitochondrial genomes. The overall base composition of the mitogenome is biased toward A + T content at 54.04%. All protein-coding genes (PCGs) began with ATG. Nine of the 13 PCGs used complete (TAA) or incomplete (TA or T) stop codon, while ND1 and ND5 ended with AGA, COI ended with AGG and ND6 with TAG. All the genes in M. ignotincta were distributed on the H-strand, except for the ND6 subunit gene and eight tRNA genes which were encoded on the L-strand.
The Timaliidae, generally known as the babblers, was a diverse family of oscine passerine birds that traditionally includes about 275 species in 50 genera (Dickinson & Christidis Citation2003). Most molecular phylogenetic work have focused on subsets of the Timaliidae (Pasquet et al. Citation2006; Zhang et al. Citation2007; Yeung et al. Citation2011). Till now, there was little information about the complete mitochondrial genomes of M. ignotincta in GenBank. In our study, the complete mitogenome of M.ignotincta was determined, which might provide some molecular data for the research on the relationships of some babblers.
Minla ignotincta (specimen voucher B43) was obtained from Ya’an, Sichuan province of China (N29.98°, E103.01°) and identified by its morphological characteristics. The complete mitochondrial genome sequence of M. ignotincta was amplified and sequenced by 18 pairs of primers with normal LA-PCR and PCR methods. The mitogenome of M. ignotincta has been deposited in Genbank (accession no. KT995474). We constructed the phylogeny by Bayesian inference (BI) using Mrbayes 3.1.1 (Huelsenbeck & Ronquist Citation2001).
Similar to other Timaliidae mitogenomes, the complete mtDNA sequence of M. ignotincta (17 868 bp in length) had 13 typical protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes (12S rRNA and 16SrRNA) and two control region (D-loop1 and D-loop2) (Zhang et al. Citation2014; Zhou et al. Citation2015). The base composition of mtDNA is 30.05% A, 23.99% T, 31.76% C and 14.20% G, so the percentage of A + T (54.04%) was slightly higher than G + C. In 13 PCGs, the shortest one was ATP8 gene (168 bp) and the longest one was the ND5 gene (1818 bp). All PCGs began with ATG. Of the 13 PCGs, nine of PCGs used complete (TAA) or incomplete (TA or T) stop codon, while ND1 and ND5 ended with AGA, COI ended with AGG and ND6 with TAG. The 12S rRNA (982 bp) and 16S rRNA (1600 bp) genes were located between the tRNAphe and tRNALeu(UUR) genes, which was separated by the tRNAVal gene. The inferred secondary structures of 21 tRNAs (excluding tRNASer(AGY)) of M. ignotincta were all conformed to the common structural features of mitochondrial tRNAs. D-loop1 and D-loop2 were located in two different locations of mitogenome. There was eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAPro and tRNAGlu) and one PCG (ND6) encoded on the L-strand, other PCGs encoded on the H-strand.
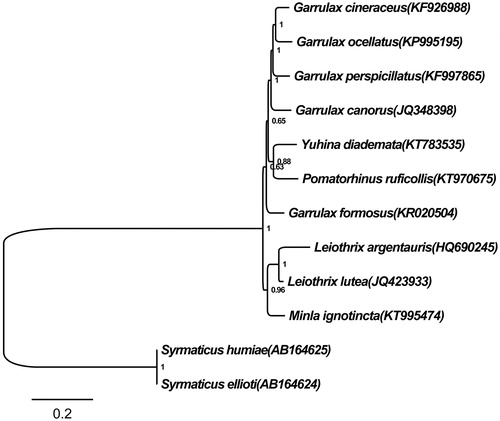
As shown in , 10 Timaliidae species clustered into two groups. A separate M. ignotincta was the sister lineage to the clade formed by the species of genus Leiothrix with more than 96% of posterior probability, which was consistent with BI analysis results of Gelang et al. (Citation2009) and Moyle et al. (Citation2012). Therefore, we hold that the genomic data of M. ignotincta was trustworthy.
Figure 1. Bayesian tree based on combining 13 protein-coding gene sequences of ten Timaliidae and two Syrmaticus birds. The Bayesian tree was reconstructed with general-time-reversible (GTR) model, and the Markov chains for 1 000 000 generations. Numbers at node of the tree branches represent posterior probability. Two Syrmaticus birds were used as the outgroup.
Disclosure statement
This work was supported by the National Natural Science Foundation of China (31370407). The authors report no conflicts of interest, and are alone responsible for the content and writing of the paper.
References
- Dickinson EC, Christidis L. 2003. The Howard & Moore complete checklist of the birds of the world, vol. 2: Passerines. 3rd ed. Eastbourne: Aves Press.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Gelang M, Cibois A, Pasquet E, Olsson U, Alström P, Ericson PGP. 2009. Phylogeny of babblers (Aves, Passeriformes): major lineages, family limits and classification. Zool Scr. 38:225–236.
- Moyle RG, Andersen MJ, Oliveros CH, Steinheimer FD, Reddy S. 2012. Phylogeny and biogeography of the core babblers (aves: timaliidae). Syst Biol. 61:631–651.
- Pasquet E, Bourdon E, Kalyakin MV, Cibois A. 2006. The fulvettas (Alcippe, Timaliidae, Aves): a polyphyletic group. Zool Scr. 35:559–566.
- Yeung CKL, Lin RC, Lei F, Robson C, Hung LM, Liang W, Zhou F, Han L, Li SH, Yang X. 2011. Beyond a morphological paradox: complicated phylogenetic relationships of the parrotbills (Paradoxornithidae, Aves). Mol Phylogenet Evol. 61:192–202.
- Zhang H, Li Y, Wu X, Xue H, Yan P, Wu X. 2016. The complete mitochondrial genome of garrulax perspicillatus (passeriformes, timaliidae). Mitochondrial DNA. 27:1265–1266.
- Zhang S, Yang L, Yang X, Yang J. 2007. Molecular phylogeny of the yuhinas (Sylviidae: Yuhina): a paraphyletic group of babblers including Zosterops and Philippine Stachyris. J Ornithol. 148:417–426.
- Zhou YY, Qi Y, Yao YF, Huan ZJ, Li, DY, Xie M, Ni QY, Zhang MW, Xu HL. 2015. Characteristic of complete mitochondrial genome and phylogenetic relationship of garrulax sannio (passeriformes, timaliidae). Mitochondrial DNA. 26:1–2.
