Abstract
Two complete mitochondrial genome sequences of ship sturgeon Acipenser nudiventris from the Caspian Sea and from Balkhash Lake (introduced from the Aral Sea during the last century) were determined by PCR-based sequencing method. The whole mitogenome sequences have been deposited in GenBank under accession numbers KU321568 and KU321569.
Ship (or Fringebarbel) sturgeon Acipenser nudiventris Lovetzky, 1828, is the only one of the Ponto-Caspian sturgeons inhabited all three previously connected marine basins – the Caspian, Black/Azov and Aral seas. This species is the most affected by recent sturgeon decline crisis. The stock in the Aral Sea was declared extinct due to catastrophic changes in water level and high salinity (Birstein Citation1993). There is no evidence for ship sturgeon occurrence in the Azov Sea for over decades, and it was not recorded in low Danube River over 30–40 years. The only three specimens have been caught in the middle part of Danube River in the last 12 years (2003 – Serbia, 2005 and 2009 – Hungary) (Danube Sturgeon Task Force website). There is a rapid decline of this species in the Caspian Sea. This indicates that Azov-and-Black Sea population is likely to be already lost and the Caspian Sea stock is on the brink of extinction. Fortunately, A. nudiventris was introduced from the Aral Sea into River Ili, Balkhash Lake basin. Total 189 adult specimens (90 in 1933 and 199 in 1934) were caught during autumn spawning migration in the Syr Darya River (5 km from the Aral Sea), transferred by specially equipped live-fish railroad car, and released into River Ili near Iliysk Railroad Station. (Dryagin Citation1953). Spawning was documented in the River Ili as early as in 1934 and 1935 and soon after ship sturgeon inhabited the whole Balkhash Lake. In the middle of 1950s, catch of the introduced species reached 20 tons annually (Dombrovsky et al. Citation1972). Later Kapchagai Dam was constructed in 1970 and separated habitat of A. nudiventris into two parts – Lake Balkhash with spawning grounds in the low part of the Ili river, and Kapchagai water reservoir with spawning grounds in the upstream part of Ili River. Both populations exist now but depressed by illegal fishing of adults and by-catch of juveniles by commercial fine-mesh fishery.
Here, we present first complete mtDNA genomes of two A. nudiventris specimens. One sample (voucher #NUD001 in Russian reference depository of sturgeon genetic samples, VNIRO, Moscow) was collected in the Caspian Sea (Ural River, Atyraubalyk Fishery Enterprise, Atyrau, Kazakhstan) in 1999. The second specimen was collected by Kazakh Fishery Institute (Almaty, Kazakhstan) in the low part of the Ili River in August 2014 and is a descendant of the extinct Aral Sea population (Voucher # NUD172). Thirty primer pairs were designed based on previously published sturgeon genomes to amplify complete genome by overlapping regions and Sanger sequenced with BigDye v3.0 chemistry from both directions by using the same primers. Primers and PCR condition presented in Supplementary Table. Assembled sequences were submitted to Genbank with accession numbers: KU321568 (voucher# NUD172, Ili River) and KU321569 (voucher#NUD001 Caspian Sea).
Mitochondrial A. nudiventris genomes from two geographically isolated populations are different by only 15 substitutions (nine A/G and six C/T), with seven of them located within the control region (CR). Multiple alignment of A. nudiventris sequences with 20 previously published complete mitochondrial genomes (Inoue et al. Citation2003; Arnason et al. Citation2004; Peng et al. Citation2007; Chen et al. Citation2012; Dong et al. Citation2014a,b; Li et al. Citation2014, Citation2015; Liao et al. Citation2016; Lu et al. Citation2016) is straightforward except CR, which is highly polymorphic and contains multiple 81-82bp repeats. Complete mtDNA sequence of A. sinensis (Genebank accession no. EU719645) was discarded from analysis because it was shown to be either incorrectly identified or an undocumented hybrid with A. gueldenstaedtii (Dillman et al. Citation2014). Bayesian phylogenetic analysis (without CR) gives single phylogenetic tree with 100% support for all branches (). Ship sturgeon clusters with A. ruthenus, as it was shown in Peng et al. (Citation2007), but A. nudiventris–A. ruthenus branch is not a sister clade to A. stellatus as it was found previously (Peng et al. Citation2007; Krieger et al. Citation2008), but has a common ancestor with high chromosome number Ponto-Caspian species such as A. gueldenstaedtii and A. baerii.
Figure 1. Bayesian tree of sturgeon species based on mitogenome sequences excluding control region. Posterior probabilities obtained in MrBayes (MrBayes Inc., River Valley, MA) are equal to 1 for all nodes.
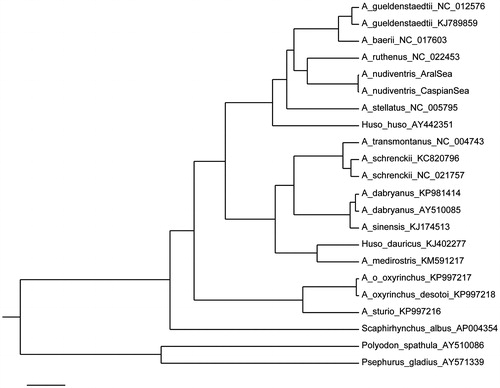
High similarity between ship sturgeon mitogenome sequences from two geographically isolated sea basins indicates that Aral and Caspian seas were connected in the recent past. Paleogeographic reconstructions propose water discharge from the Great West Siberia glacial lake to the Aral Sea and further to the Caspian Sea (Mangerud et al. Citation2004), and, therefore, the Caspian and Aral ship sturgeon populations were connected about 90 kya.
Mugue_Supplemental_material.doc
Download ()Disclosure statement
The authors report no conflict of interest. This present research was supported by grant from Russian Foundation of Basic Research (RFBR#15-28-02766). The authors alone are responsible for the content and writing of the paper.
References
- Arnason U, Gullberg A, Janke A, Joss J, Elmerot C. 2004. Mitogenomic analyses of deep gnathostome divergences: a fish is a fish. Gene. 333:61–70.
- BirsteinVJ. 1993. Sturgeons and paddle fishes: threatened fishes in need of conservation. Conserv Biol. 7:773–787.
- Birstein VJ, Doukakis P, Desalle R. 2002. Molecular phylogeny of acipenseridae: nonmonophyly of scaphirhynchinae. Copeia. 2002:287–301.
- Chen XW, Jiang S, Shi ZY, Li Q, Xun XR, Guo Da Q. 2012. Mitochondrial genome of the Siberian sturgeon Acipenserbaerii. Mitochondrial DNA. 23:120–122.
- Danube Sturgeon Task Force webpage; [cited 2015 Dec 27]. Available from: http://www.dstf.eu/species/acipenser-nudiventris/.
- Dillman CB, Zhuang P, Zhang T, Zhang LZ, Mugue N, Hilton EJ. 2014. Forensic investigations in to a Gen Bank anomaly: endangered taxa and the importance of voucher specimens in molecular studies. J Appl Ichthyol. 30:1300–1309.
- Dombrovsky GV, Serov NP, Dikanskiy VN. 1972. Biology and fishery of ship sturgeon Acipenser nudiventris Lov. In the Balkhash-Ilibasin. Proc. TzNIORKh 4:146–148. [InRussian].
- Dryagin PA. 1953. Fish acclimatization in the interior water bodies of the USSR. Izvestia VNIORKh. 32:10–98. [In Russian].
- Dong C, Chen B, Xu J, Mahboob S, Al-Ghanim K, Xu P, Sun X. 2014. The complete mitochondrial genome of Russian sturgeon (Acipensergueldenstaedti). Mitochondrial DNA. 27:1–2.
- Dong XL, Yu J, Zhang Y, Sun DJ. 2014. Complete mitochondrial genome of Acipenserschrenckii (Acipenseriformes, Acipenseridae). Mitochondrial DNA. 25:350.
- Inoue JG, Miya M, Tsukamoto K, Nishida M. 2003. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the “ancient fish”. Mol Phylogenet Evol. 26:110–120.
- Krieger J, Hett AK, Fuerst PA, Artyukhin EA, Ludwig A. 2008. The molecular phylogeny of the order Acipenseriformes revised. J Appl Ichthyol. 24:36–45.
- Li C, Lu CY, Li JT, Cheng L, Zheng XH, Sun XW. 2014. Complete mitochondrial genome of Amursturgeon (Acipenserschrenckii). Mitochondrial DNA. 25:282–283.
- Liao X, Tian H, Zhu B, Chang J. 2016. The complete mitochondrial genome of Chinese sturgeon (Acipensersinensis). Mitochondrial DNA. 27:328–329.
- Li C, Cheng L, Li JT, Lu CY, Wang Y, Sun XW. 2015. Complete mitochondrial genome of sterlet (Acipenserruthenus). Mitochondrial DNA. 26:259–260.
- Lu C, Gu Y, Li C, Cheng L, Sun X. 2016. The complete mitochondrial genome of Endangered fish Husodauricus (Acipenseriformes: Acipenseridae). Mitochondrial DNA. 27:395–396.
- Mangerud J, Jakobsson M, Alexandersonk H, Astakhov V, Clarkee GKC, Henriksena M, Hjortc C, Krinnerf G, Lunkka J-P, Moller P, et al. 2004. Ice-dammed lakes and rerouting of the drainage of northern Eurasia during the Last Glaciation. Quat Sci Rev. 23:1313–1332.
- Peng Z, Ludwig A, Wang DQ, Diogo R, Wei Q, He S. 2007. Age and biogeography of major clades in sturgeons and paddle fishes (Pisces: Acipenseriformes). Mol Phylogenet Evol. 42:854–862.
- Sturgeon Specialist Group. 1996. Acipenser nudiventris (Aral Sea stock). The IUCN Red List of Threatened Species 1996:e.T251A13048150; [cited 2015 Dec 27]. Available from: http://dx.doi.org/10.2305/IUCN.UK.1996.RLTS.T251A13048150.en.
