Abstract
In this study, we report the complete mitochondrial genome sequence of Achatinella mustelina, an endangered Hawaiian tree snail. The mitogenome is 16 323 bp in length and has a base composition of A (34.7%), T (42.6%), C (12.7%) and G (10.0%). Similar to other Pulmonates, it contains 13 protein-coding genes, 2 rRNA genes and 22 tRNA genes. To our knowledge, this is the first mitochondrial genome sequenced within the Achatinelloidea superfamily, which contains a high number of endangered species. As such, this mitogenome will be useful in conservation genetics studies.
The genus Achatinella, endemic to the island of O‘ahu, included at least 41 species of Hawaiian tree snails (Pilsbry & Cooke Citation1912–1914). Habitat loss, predation by introduced species and over-harvesting by collectors led to the extinction of at least 30 species of Achatinella, and resulted in the declaration of all remaining species in the genus as Endangered (Hadfield & Mountain Citation1980; U.S.A Fish and Wildlife Service Citation1981; Hadfield Citation1986). Of these, Achatinella mustelina is the most abundant, with at least 2000 individuals remaining in the wild.
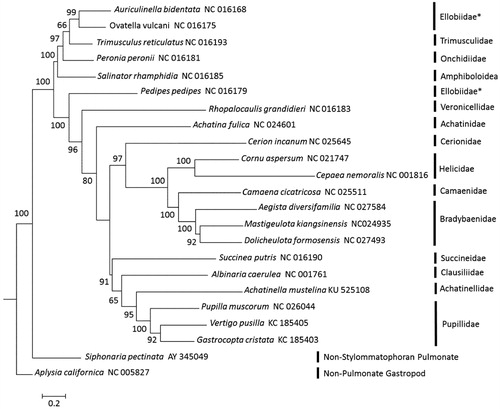
Figure 1. Placement of Achatinella mustelina among Stylommatophoran snails. Alignments, model tests and maximum likelihood analyses were performed using MEGA version 6.2 (Tamura et al. Citation2013). The 13 protein-coding mitochondrial gene sequences were individually translated into amino acid sequences, and then aligned using ClustalW in MEGA version 6.2 (Tamura et al. Citation2013). Default settings were used with the following exception: the multiple alignment parameters were changed to a gap opening penalty of 3.0; and the gap extension penalty was set to 1.8. The amino acid substitution model was found to be LG + G + I + F using the Akaike Information Criterion (AIC). Maximum likelihood analysis of the amino acid sequences was run using the identified model, with bootstrap support values based on 1000 replicates. The resulting tree shows similar relationships to previous studies, though species in one family (Ellobiidae) did not group together (Hatzoglou et al. Citation1995; Grande et al. Citation2004; Knudsen et al. Citation2006; White et al. Citation2011; Yamazaki et al. Citation1997; Gaitán-Espitia et al. Citation2013; Gonzalez et al. 2014 (unpublished); He et al. Citation2014; Wang et al. Citation2014; Huang et al. Citation2015; Marquardt et al. 2015 (unpublished); Deng et al. Citation2016).
We sequenced the complete mitochondrial genome of A. mustelina (GenBank accession number KU525108). Small tissue samples were collected from 15 populations across the range (15–40 individuals per population), using non-lethal methods, and preserved in 100% ethanol until DNA extraction (Thacker & Hadfield Citation2000). DNA was individually extracted from tissue samples using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Extracted DNA was quantified using the Biotium AccuClear Ultra High Sensitivity dsDNA quantitation kit, Hayward, CA with seven standards. Equal quantities of DNA from each individual within a population were pooled to a total of 1μg. From these pools, libraries were prepared for genome scanning using the ezRAD protocol (Toonen et al. Citation2013) version 2.0 (http://ingridknapp.weebly.com/current-research.html). Samples were digested with the frequent cutter restriction enzyme DpnII from New England Biolabs®, Ipswich, MA and prepared for sequencing on the Illumina® MiSeq using the Kapa Biosystems Hyper Prep kit, Wilmington, MA. All samples were amplified to generate 1 μg of adapter-ligated DNA, then validated and quantified to ensure equal pooling on the MiSeq flow cell, using a Bioanalyzer and qPCR. Quality control checks and sequencing were performed by the Hawaii Institute of Marine Biology Genetics Core Facility.
Reads were cleaned, and then assembled using an iterative method within Geneious 6.0, Newark, NJ. Ends were trimmed to remove barcode sequences, adapters, primers and low-quality regions (quality score <20). We obtained 69 178 116 sequences (mean ± S.D. = 4 494 912 ± 1 818 900 per population).
Reads were initially mapped to the mitogenome of Albinuria coerulea (Hatzoglou et al. Citation1995). The alignment of mapped sequences was inspected, and a consensus sequence was generated. This consensus sequence was used as a reference for the next iteration, in which all ∼69 million sequences were mapped against the consensus sequence achieved in the previous round of alignment. This process was repeated until the complete mitochondrial genome was obtained. In total, 30 695 reads mapped to the complete mitochondrial genome, with coverage ranging from 10× to 4409× per site (256 ± 50). Annotation of mitochondrial elements was carried out with DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013).
The Achatinella mustelina mitogenome is similar to those of other Pulmonates (), with 13 protein-coding genes, 2 rRNA genes and 22 tRNA genes. The total length is 16 323 bp, slightly larger than other Pulmonates (White et al. Citation2011). The base composition of the genome is: A (34.7%), T (42.6%), C (12.7%) and G (10.0%). This is the first mitochondrial genome sequenced within the Achatinelloidea superfamily.
Collection sites
Samples were collected from the following locations (decimal degrees): 21.413406, -158.099816; 21.438595, -158.097806; 21.467780, -158.103860; 21.476095, -158.125562; 21.488028, -158.138127; 21.498169, -158.143955; 21.505067, -158.127069; 21.510198, -158134197; 21.514961, -158153266; 21.525394, -158.170789; 21.496982, -158.172874; 21.511052, -158.190455; 21.514602, -158.201006; 21.537079, -158.199123; 21.538785, -158.198855.
Funding information
This project was funded by the Oahu Army Natural Resources Program through the University of Alaska, Anchorage, under grant agreement 2911KB-14-2-0001. Funding was provided by the Oahu Army Natural Resources Program through the University of Alaska, Anchorage.
Acknowledgements
We appreciate the invaluable field assistance provided by V Costello, J Tanino, D Sischo and J Prior.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Bernt M, Donath A, Jüling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Deng PJ, Wang WM, Huang XC, Wu XP, Xie GL, Ouyang S. 2016. The complete mitochondrial genome of Chinese land snail Mastigeulota kiangsinensis (Gastropoda: Pulmonata: Bradybaenidae). Mitochondrial DNA. 27:1441–1442.
- Gaitán-Espitia JD, Nespolo RF, Opazo JC. 2013. The complete mitochondrial genome of the land snail Cornu aspersum (Helicidae: Mollusca): intra-specific divergence of protein-coding genes and phylogenetic considerations within Euthyneura. PLoS One. 8:e67299.
- Grande C, Templado J, Cervera JL, Zardoya R. 2004. Molecular phylogeny of euthyneura (mollusca: gastropoda). Mol Biol Evol. 21:303–313.
- Hadfield MG. 1986. Extinction in Hawaiian Achatinelline snails. Malacologia. 27:67–81.
- Hadfield MG, Mountain BS. 1980. A field study of a vanishing species, Achatinella mustelina (Gastropoda, Pulmonata), in the Waianae Mountains of Oahu. Pac Sci. 34:345–358.
- Hatzoglou E, Rodakis GC, Lecanidou R. 1995. Complete sequence and gene organization of the mitochondrial genome of the land snail Albinaria coerulea. Genetics. 140:1353–1366.
- He ZP, Dai XB, Zhang S, Zhi TT, Lun ZR, Wu ZD, Yang TB. 2014. Complete mitochondrial genome of the giant African snail, Achatina fulica (Mollusca: Achatinidae): a novel location of putitive control regions (CR) in the mitogenome within Pulmonate species. Mitochondrial DNA. 27:1084–1085.
- Huang CW, Lin SM, Wu WL. 2015. Mitochondrial genome sequences of landsnails Aegista diversifamilia and Dolicheulota formosensis (Gastropoda: Pulmonata: Stylommatophora). Mitochondrial DNA. 22:1–3.
- Knudsen B, Kohn AB, Nahir B, McFadden CS, Moroz LL. 2006. Complete DNA sequence of the mitochondrial genome of the sea-slug, Aplysia californica: conservation of the gene order in Euthyneura. Mol Phylogenet Evol. 38:459–469.
- Pilsbry HA, Cooke CM. 1912–1914. Achatinellidae: Manual of Conchology, 2nd ser. Vol. 21 lviii + 428 pages 63 plates.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2729–3735.
- Thacker RW, Hadfield MG. 2000. Mitochondrial phylogeny of extant Hawaiian tree snails (Achatinellinae). Mol. Phylogenet. Evol. 16:263–270.
- Toonen R J, Puritz J B, Forsman Z H, Whitney J, Fernandez-Silva I, Andrews K, Bird C E. 2013. ezRAD: a simplified method for genomic genotyping in non-model organisms. Peer J. 1:e203 http://dx.doi.org/10.7717/peerj.203.
- U.S. Fish and Wildlife Service. 2013. Endangered and threatened wildlife and plants; determination of endangered status for 38 species on Molokai, Lanai, and Maui. Federal Register. 78:32014–32065.
- Wang P, Yang HF, Zhou WC, Hwang CC, Zhang WH, Qian ZX. 2014. The mitochondrial genome of the land snail Camaena cicatricosa (Müller, 1774): the first complete sequence in the family Camaenidae. Zookeys. 451:33–48.
- White TR, Conrad MM, Tseng R, Balayan S, Golding R, de Frias Martins AM, Dayrat BA. 2011. Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetic relationships. BMC Evol Biol. 11:295.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yamazaki N, Ueshima R, Terrett JA, Yokobori S, Kaifu M, Segawa R, Kobayashi T, Numachi K, Ueda T, Nishikawa K, Watanabe K, Thomas RH. 1997. Evolution of pulmonate gastropod mitochondrial genomes: Comparisons of gene organizations of Euhadra, Cepaea and Albinaria and implications of unusual tRNA secondary structures. Genetics. 145:749–758.
