Abstract
We have sequenced the complete mitochondrial genome (mitogenome) of the pine moth Dendrolimus spectabilis Butler (Lepidoptera: Lasiocampidae), which has been a serious pest for the Japanese red pine (Pinus densiflora S. et Z.) in Korea. The 15 409 bp complete mitochondrial genome (mitogenome) of the species consists of a typical set of genes (13 protein-coding genes, 2 rRNA genes and 22 tRNA genes) and the A + T-rich region, with an arrangement typical of Ditrysia in Lepidoptera. The 320 bp long A + T-rich region of D. spectabilis contains the motif ATAGA near the 5′-end of the srRNA, with a 14 bp-long poly-T stretch – the ATTTA sequence – and a microsatellite-like sequence consisting of (TA)7, but lacks the poly-A stretch often found immediately upstream of tRNAMet in other Lepidoptera. A phylogenetic analysis using 13 protein-coding genes from Bombycoidea and Lasiocampoidea has shown that D. spectabilis was placed as a sister to the congeneric species group, D. tabulaeformis + D. punctatus.
The pine moth Dendrolimus spectabilis Butler (Lepidoptera: Lasiocampidae) is distributed in Korea, Japan, China and Russia, and had been a serious pest for the Japanese red pine (Pinus densiflora S. et Z.) during the 1960s and 1970s (Shin et al. Citation2008). Analysis of the species distribution in Korea has shown that the phenology in Korea has changed from univoltinism to biovoltinism owing to climate change (Choi et al. Citation2011), although this information is not available for other regions. In order to obtain genomic information for the species, we sequenced the complete mitochondrial genome (mitogenome) of D. spectabilis. One adult was captured from a mountainous area at Uido Island, Sinan-gun, Jeollanam-do Province, South Korea (34°36′11″ N, 125°52′14″ E). A voucher specimen was deposited in Chonnam National University, Gwangju, Korea.
The complete mitogenome was amplified into three overlapping long fragments, using genomic DNA as a template, and subsequently into 26 overlapping short fragments, using the long fragments as templates. The primers were adapted from those used by Kim et al. (Citation2012).
The D. spectabilis mitogenome is 15 409 bp in length and includes typical sets of genes (2 rRNAs, 22 tRNAs and 13 protein-coding genes) and a major non-coding 320 bp A + T-rich region (GenBank accession number KU558688). The size of the total genome and of the A + T-rich region is nearly identical to those of congeneric species, including that of the same species as reported from China (Qin et al. Citation2015), indicating that the change in voltinism does not appear to be accompanied by changes in general genetic constitution. Gene arrangement in D. spectabilis was identical to that of the major lepidopteran group, Ditrysia, in Lepidoptera, which is ordered tRNAMet/tRNAIle/tRNAGln between the A + T-rich region and ND2 (Kim et al. Citation2011). Twelve of the 13 PCGs began with typical ATN codons, but the COI began with CGA (data not shown), which is highly conserved in Lepidoptera (Kim et al. Citation2014).
The 320-bp long A + T-rich region of D. spectabilis contains typical conserved sequences, such as the motif ATAGA close to a 5′-end of the srRNA, with a 14-bp long poly-T stretch. This motif and the poly-T stretch have been suggested to be the site of replication origin of the minority strand for mtDNA in the lepidopteran Bombyx mori (Saito et al. Citation2005). Along with this motif, the A + T-rich region of D. spectabilis contains the ATTTA sequence, the function of which is unknown, and a microsatellite-like sequence consisting of (TA)7. However, the A + T-rich region of D. spectabilis lacks a poly-A stretch, which is often found immediately upstream of tRNAMet in other lepidopteran mitogenomes (Kim et al. Citation2014).
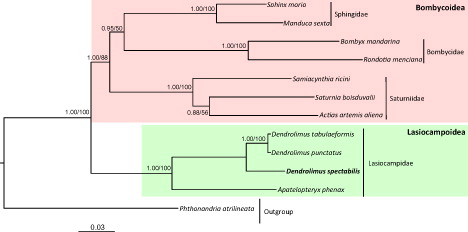
We downloaded the available mitogenome sequences of 10 Bombycoidea and Lasiocampoidea species, along with one species of Geometroidea for use as an outgroup, and 13 PCGs were utilized for phylogenetic analysis. Maximum-likelihood (ML) and Bayesian inference (BI) methods were performed using the GTR + I + G model in CIPRES Portal v. 3.1 (La Jolla, CA; Miller et al. Citation2010). The two analytical methods presented an identical topology, showing the monopoly of each superfamily with high nodal supports and indicating that D. spectabilis is a sister to the congeneric species group, D. tabulaeformis + D. punctatus, rendering Dendrolimus a strong monophyletic group (BI, 1.0 and ML, 100%; ).
Figure 1. Phylogeny of Lasiocampoidea and Bombycoidea. Bayesian Inference (BI) and Maximum Likelihood (ML) methods produced the same topology based on concatenated 13 PCGs. The numbers at each node specify Bayesian posterior probabilities as percentages calculated by BI analysis (first value) and bootstrap percentages of 1000 pseudoreplicates by ML analysis (second value). The scale bar indicates the number of substitutions per site. One species of Geometroidea was utilized as an outgroup. GenBank accession numbers are as follows: Sphinx morio, KC470083; Manduca sexta, EU286785; Bombyx mandarina, AB070263; Rondotia menciana, KJ647172; Actias artemis aliena, KF927042; Samia cynthia ricini, JN215366; Saturnia boisduvalii, EF622227; Dendrolimus tabulaeformis, KJ913817; Dendrolimus punctatus, KJ913813; Apatelopteryx phenax, KJ508055 and Phthonandria atrilineata, EU569764.
Funding information
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2015R1D1A3A03018119).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Choi WI, Park Y-K, Park Y-S, Ryoo MI, Lee H-P. 2011. Changes in voltinism in a pine moth Dendrolimus spectabilis (Lepidoptera: Lasiocampidae) population: implications of climate change. Appl Entomol Zool. 46:319–325.
- Kim MJ, Kang AR, Jeong HC, Kim K-G, Kim I. 2011. Reconstructing intraordinal relationships in Lepidoptera using mitochondrial genome data with the description of two newly sequenced lycaenids, Spindasis takanonis and Protantigius superans (Lepidoptera: Lycaenidae). Mol Phylogenet Evol. 61:436–445.
- Kim JS, Park JS, Kim MJ, Kang PD, Kim SG, Jin BR, Han YS, Kim I. 2012. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia cynthia ricini (Lepidoptera: Saturniidae). J Asia Pac Entomol. 15:162–173.
- Kim MJ, Wang AR, Park JS, Kim I. 2014. Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene. 549:97–112.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of Gateway Comput Environ Workshop (GCE), New Orleans, pp. 1–8.
- Qin J, Zhang Y, Zhou X, Kong X, Wei S, Ward RD, Zhang A. 2015. Mitochondrial phylogenomics and genetic relationships of closely related pine moth (Lasiocampidae: Dendrolimus) species in China, using whole mitochondrial genomes. BMC Genomics. 16:428–39.
- Saito S, Tamura K, Aotsuka T. 2005. Replication origin of mitochondrial DNA in insects. Genetics. 171:1695–1705.
- Shin SC, Choi KS, Choi WI, Chung YJ, Lee SG, Kim CS. 2008. A new illustrated book of forest insect pests. Seoul: Upgo MunHwa.
